bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2728_orf2
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
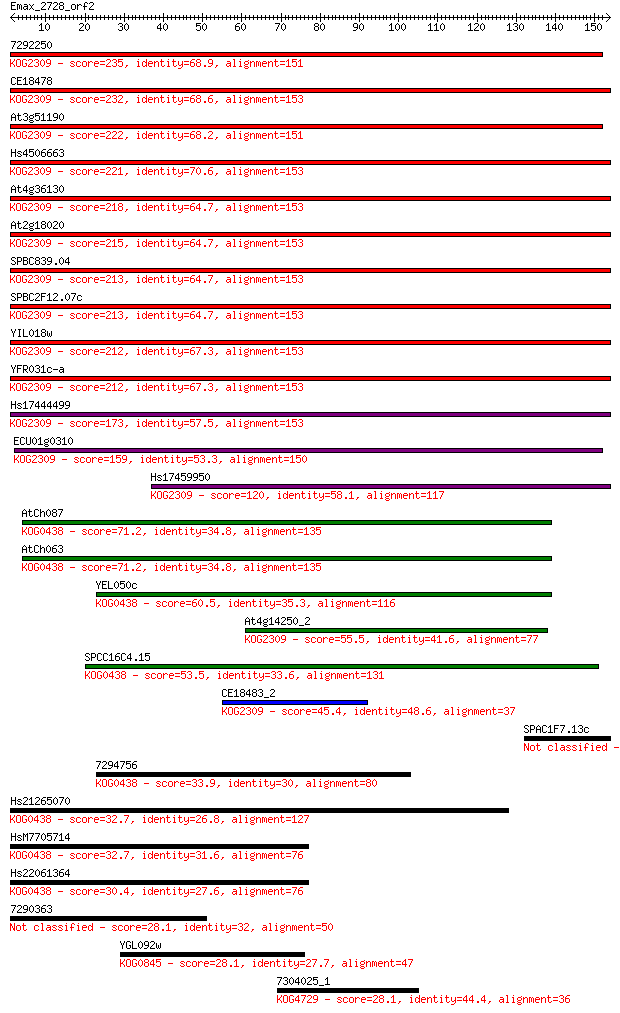
7292250 235 3e-62
CE18478 232 2e-61
At3g51190 222 2e-58
Hs4506663 221 4e-58
At4g36130 218 3e-57
At2g18020 215 3e-56
SPBC839.04 213 9e-56
SPBC2F12.07c 213 9e-56
YIL018w 212 2e-55
YFR031c-a 212 2e-55
Hs17444499 173 1e-43
ECU01g0310 159 1e-39
Hs17459950 120 7e-28
AtCh087 71.2 7e-13
AtCh063 71.2 7e-13
YEL050c 60.5 1e-09
At4g14250_2 55.5 4e-08
SPCC16C4.15 53.5 2e-07
CE18483_2 45.4 4e-05
SPAC1F7.13c 38.1 0.006
7294756 33.9 0.14
Hs21265070 32.7 0.25
HsM7705714 32.7 0.27
Hs22061364 30.4 1.4
7290363 28.1 6.2
YGL092w 28.1 6.6
7304025_1 28.1 7.0
> 7292250
Length=256
Score = 235 bits (599), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 104/151 (68%), Positives = 133/151 (88%), Gaps = 0/151 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQF++CG+KA L IGNV+PL ++PEG+++C++EEK GDRG LA+TSG++ATV
Sbjct 77 IAPEGMHTGQFVYCGRKATLQIGNVMPLSQMPEGTIICNLEEKTGDRGRLARTSGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+++T K+RV+LPSGA+K +PS +R ++G+VAGGGRIDKP+LKAG AYHKYK KRN W
Sbjct 137 IAHNQDTKKTRVKLPSGAKKVVPSANRAMVGIVAGGGRIDKPILKAGRAYHKYKVKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSR 151
KVRGVAMNPVEHPHGGGNHQHIG STV R
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGKASTVKR 227
> CE18478
Length=260
Score = 232 bits (591), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 105/153 (68%), Positives = 130/153 (84%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFIHCG KA + IGN++P+G +PEG+ +C+VE K GDRG +A+ SG++ATV
Sbjct 77 VAAEGMHTGQFIHCGAKAQIQIGNIVPVGTLPEGTTICNVENKSGDRGVIARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ +T K+R+RLPSGA+K + S +R +IGLVAGGGR DKPLLKAG +YHKYKAKRN W
Sbjct 137 IAHNPDTKKTRIRLPSGAKKVVQSVNRAMIGLVAGGGRTDKPLLKAGRSYHKYKAKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHPHGGGNHQHIGHPSTV R A
Sbjct 197 PRVRGVAMNPVEHPHGGGNHQHIGHPSTVRRDA 229
> At3g51190
Length=260
Score = 222 bits (566), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 103/151 (68%), Positives = 125/151 (82%), Gaps = 0/151 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQ+++CGKKA L +GNVLPLG +PEG+V+C+VE GDRGALA+ SG +A V
Sbjct 78 VAAEGMYTGQYLYCGKKANLMVGNVLPLGSIPEGAVICNVELHVGDRGALARASGDYAIV 137
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ E+ +RV+LPSG++K LPS R +IG VAGGGR +KP LKAG AYHKYKAKRNCW
Sbjct 138 IAHNPESNTTRVKLPSGSKKILPSACRAMIGQVAGGGRTEKPFLKAGNAYHKYKAKRNCW 197
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSR 151
VRGVAMNPVEHPHGGGNHQHIGH STV R
Sbjct 198 PVVRGVAMNPVEHPHGGGNHQHIGHASTVRR 228
> Hs4506663
Length=257
Score = 221 bits (563), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 108/153 (70%), Positives = 134/153 (87%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIHTGQF++CGKKA L+IGNVLP+G +PEG++VC +EEKPGDRG LA+ SG++ATV
Sbjct 77 IAAEGIHTGQFVYCGKKAQLNIGNVLPVGTMPEGTIVCCLEEKPGDRGKLARASGNYATV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET K+RV+LPSG++K + S +R ++G+VAGGGRIDKP+LKAG AYHKYKAKRNCW
Sbjct 137 ISHNPETKKTRVKLPSGSKKVISSANRAVVGVVAGGGRIDKPILKAGRAYHKYKAKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMNPVEHP GGGNHQHIG PST+ R A
Sbjct 197 PRVRGVAMNPVEHPFGGGNHQHIGKPSTIRRDA 229
> At4g36130
Length=258
Score = 218 bits (555), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 123/153 (80%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGKKA L +GNVLPL +PEG+V+C+VE GDRG A+ SG +A V
Sbjct 77 VAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVICNVEHHVGDRGVFARASGDYAIV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ + SR++LPSG++K +PS R +IG VAGGGR +KP+LKAG AYHKY+ KRNCW
Sbjct 137 IAHNPDNDTSRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNCW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGH STV R A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDA 229
> At2g18020
Length=258
Score = 215 bits (547), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 124/153 (81%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGKKA L +GNVLPL +PEG+VVC+VE GDRG LA+ SG +A V
Sbjct 77 VAAEGMYTGQFLYCGKKATLVVGNVLPLRSIPEGAVVCNVEHHVGDRGVLARASGDYAIV 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ++ +R++LPSG++K +PS R +IG VAGGGR +KP+LKAG AYHKY+ KRN W
Sbjct 137 IAHNPDSDTTRIKLPSGSKKIVPSGCRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
KVRGVAMNPVEHPHGGGNHQHIGH STV R A
Sbjct 197 PKVRGVAMNPVEHPHGGGNHQHIGHASTVRRDA 229
> SPBC839.04
Length=253
Score = 213 bits (543), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 135/153 (88%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGK AAL++GNVLP+G++PEG+++ +VEEK GDRGAL ++SG++ +
Sbjct 76 VATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKAGDRGALGRSSGNYVII 135
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
VGH +TGK+RV+LPSGA+K +PS++R ++G+VAGGGRIDKPLLKAG A+HKY+ KRNCW
Sbjct 136 VGHDVDTGKTRVKLPSGAKKVVPSSARGVVGIVAGGGRIDKPLLKAGRAFHKYRVKRNCW 195
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+ RGVAMNPV+HPHGGGNHQH+GH +TV R +
Sbjct 196 PRTRGVAMNPVDHPHGGGNHQHVGHSTTVPRQS 228
> SPBC2F12.07c
Length=253
Score = 213 bits (543), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 135/153 (88%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG++TGQF++CGK AAL++GNVLP+G++PEG+++ +VEEK GDRGAL ++SG++ +
Sbjct 76 VATEGMYTGQFVYCGKNAALTVGNVLPVGEMPEGTIISNVEEKAGDRGALGRSSGNYVII 135
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
VGH +TGK+RV+LPSGA+K +PS++R ++G+VAGGGRIDKPLLKAG A+HKY+ KRNCW
Sbjct 136 VGHDVDTGKTRVKLPSGAKKVVPSSARGVVGIVAGGGRIDKPLLKAGRAFHKYRVKRNCW 195
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+ RGVAMNPV+HPHGGGNHQH+GH +TV R +
Sbjct 196 PRTRGVAMNPVDHPHGGGNHQHVGHSTTVPRQS 228
> YIL018w
Length=254
Score = 212 bits (540), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 103/153 (67%), Positives = 132/153 (86%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFI+ GKKA+L++GNVLPLG VPEG++V +VEEKPGDRGALA+ SG++ +
Sbjct 77 IANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDRGALARASGNYVII 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+GH+ + K+RVRLPSGA+K + S++R +IG++AGGGR+DKPLLKAG A+HKY+ KRN W
Sbjct 137 IGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLLKAGRAFHKYRLKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
K RGVAMNPV+HPHGGGNHQHIG ST+SR A
Sbjct 197 PKTRGVAMNPVDHPHGGGNHQHIGKASTISRGA 229
> YFR031c-a
Length=254
Score = 212 bits (540), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 103/153 (67%), Positives = 132/153 (86%), Gaps = 0/153 (0%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EG+HTGQFI+ GKKA+L++GNVLPLG VPEG++V +VEEKPGDRGALA+ SG++ +
Sbjct 77 IANEGVHTGQFIYAGKKASLNVGNVLPLGSVPEGTIVSNVEEKPGDRGALARASGNYVII 136
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+GH+ + K+RVRLPSGA+K + S++R +IG++AGGGR+DKPLLKAG A+HKY+ KRN W
Sbjct 137 IGHNPDENKTRVRLPSGAKKVISSDARGVIGVIAGGGRVDKPLLKAGRAFHKYRLKRNSW 196
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
K RGVAMNPV+HPHGGGNHQHIG ST+SR A
Sbjct 197 PKTRGVAMNPVDHPHGGGNHQHIGKASTISRGA 229
> Hs17444499
Length=239
Score = 173 bits (438), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 88/153 (57%), Positives = 110/153 (71%), Gaps = 5/153 (3%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATV 60
+A EGIH GQF++CGKK L+IG+VL + +PE + VC ++EKP DRG LA+ SG+ ATV
Sbjct 64 IAAEGIHMGQFVYCGKKVQLNIGSVLSVDTMPESTTVCCLKEKPEDRGKLARASGNGATV 123
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ H+ ET K+ A +R + G+VA GGR DK +LKAG A+HKYKAKRNCW
Sbjct 124 ISHNPETQKTHE-----AALLRQQANRAVGGVVARGGRNDKAILKAGRAHHKYKAKRNCW 178
Query 121 HKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
+VRGVAMN VEHP GGGNHQHIG PST+ R A
Sbjct 179 PRVRGVAMNLVEHPFGGGNHQHIGKPSTIRRDA 211
> ECU01g0310
Length=239
Score = 159 bits (403), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 80/150 (53%), Positives = 107/150 (71%), Gaps = 0/150 (0%)
Query 2 AVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVV 61
A EG++TGQ I G A + IGN+ + VPEG V SVE GD G A +GS++ VV
Sbjct 64 ASEGMYTGQKILIGDDAPIDIGNITKIKNVPEGMAVNSVESVYGDGGTFAMVNGSYSLVV 123
Query 62 GHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH 121
H +ET ++ +++PSG + T+ S RCI+G+VAGGG DKPLLKA A++K KA+ + +
Sbjct 124 NHRKETNETVIKIPSGKKLTVSSECRCIVGVVAGGGIHDKPLLKASVAHYKAKARGHVFP 183
Query 122 KVRGVAMNPVEHPHGGGNHQHIGHPSTVSR 151
+VRGVAMNPVEH HGGGNHQH+G P+TVS+
Sbjct 184 RVRGVAMNPVEHIHGGGNHQHVGKPTTVSK 213
> Hs17459950
Length=124
Score = 120 bits (302), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 68/117 (58%), Positives = 88/117 (75%), Gaps = 9/117 (7%)
Query 37 VCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGG 96
+C +EEKP D G L + SG++ATV+ H+ ET KSRV+LPSG++K S +R ++G+VAGG
Sbjct 1 MCCLEEKPRDHGKLTQASGNYATVISHNPETKKSRVKLPSGSKKVTSSANRAVVGVVAGG 60
Query 97 GRIDKPLLKAGTAYHKYKAKRNCWHKVRGVAMNPVEHPHGGGNHQHIGHPSTVSRHA 153
G+IDKP+LKAG AYH+YKAKRNCW +VRG GGG+HQHIG PST+ R A
Sbjct 61 GQIDKPILKAGWAYHRYKAKRNCWPQVRG---------GGGGSHQHIGKPSTICRDA 108
> AtCh087
Length=274
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 71/139 (51%), Gaps = 15/139 (10%)
Query 4 EGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGH 63
G G I G + + +GN LPL +P G+ + ++E G G LA+ +G+ A ++
Sbjct 106 RGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI-- 163
Query 64 SEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH-- 121
++E + ++LPSG + + N +G V G K L +AG+ CW
Sbjct 164 AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGK 214
Query 122 --KVRGVAMNPVEHPHGGG 138
VRGV MNPV+HPHGGG
Sbjct 215 RPVVRGVVMNPVDHPHGGG 233
> AtCh063
Length=274
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 71/139 (51%), Gaps = 15/139 (10%)
Query 4 EGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGH 63
G G I G + + +GN LPL +P G+ + ++E G G LA+ +G+ A ++
Sbjct 106 RGAIIGDTIVSGTEVPIKMGNALPLTDMPLGTAIHNIEITLGRGGQLARAAGAVAKLI-- 163
Query 64 SEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH-- 121
++E + ++LPSG + + N +G V G K L +AG+ CW
Sbjct 164 AKEGKSATLKLPSGEVRLISKNCSATVGQVGNVGVNQKSLGRAGS---------KCWLGK 214
Query 122 --KVRGVAMNPVEHPHGGG 138
VRGV MNPV+HPHGGG
Sbjct 215 RPVVRGVVMNPVDHPHGGG 233
> YEL050c
Length=393
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/120 (34%), Positives = 63/120 (52%), Gaps = 13/120 (10%)
Query 23 GNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKTL 82
GN LP+ +P G+++ +V P G +++G++A V+ E K+ VRL SG + +
Sbjct 238 GNCLPISMIPIGTIIHNVGITPVGPGKFCRSAGTYARVLAKLPEKKKAIVRLQSGEHRYV 297
Query 83 PSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH----KVRGVAMNPVEHPHGGG 138
+ IG+V+ ++ L KAG R+ W VRGVAMN +HPHGGG
Sbjct 298 SLEAVATIGVVSNIDHQNRSLGKAG---------RSRWLGIRPTVRGVAMNKCDHPHGGG 348
> At4g14250_2
Length=67
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 43/78 (55%), Gaps = 12/78 (15%)
Query 61 VGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCW 120
+ +S + + LP ++KT+ S R +IG +A G K ++K RN W
Sbjct 1 IANSMISVTWDINLPLDSKKTVLSGCRVMIGQIASSGLTKKLMIK-----------RNMW 49
Query 121 HKVRGVAM-NPVEHPHGG 137
KVRGVAM NPVEHPHGG
Sbjct 50 AKVRGVAMMNPVEHPHGG 67
> SPCC16C4.15
Length=318
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/135 (32%), Positives = 67/135 (49%), Gaps = 15/135 (11%)
Query 20 LSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGAR 79
L GN PL +P G+V+ ++ P + L +++GS A ++ + + VRL SG
Sbjct 162 LKPGNCFPLRLIPIGTVIHAIGVNPNQKAKLCRSAGSSARIIAFDGKY--AIVRLQSGEE 219
Query 80 KTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKAKRNCWH----KVRGVAMNPVEHPH 135
+ + S IG+V+ + L KAG R+ W VRG AMNP +HPH
Sbjct 220 RKILDTSFATIGVVSNIYWQHRQLGKAG---------RSRWLGIRPTVRGTAMNPCDHPH 270
Query 136 GGGNHQHIGHPSTVS 150
GGG + IG+ + S
Sbjct 271 GGGGGKSIGNKPSQS 285
> CE18483_2
Length=72
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 55 GSFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIG 91
G++A V+ H+ +T K+R+RLPSGA+K + S +R +IG
Sbjct 1 GNYANVIAHNADTKKTRIRLPSGAKKVVQSVNRAMIG 37
> SPAC1F7.13c
Length=47
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 19/22 (86%), Gaps = 0/22 (0%)
Query 132 EHPHGGGNHQHIGHPSTVSRHA 153
+HPHGGGNHQH+GH +TV R +
Sbjct 1 DHPHGGGNHQHVGHSTTVPRQS 22
> 7294756
Length=294
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 8/80 (10%)
Query 23 GNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLPSGARKTL 82
G+ PLG +P G+ + +E+ PG L +G+F T++ +E K V+LPS ++
Sbjct 161 GDAYPLGALPVGTRIHCLEKNPGQMCHLIHAAGTFGTILRKFDE--KVVVQLPS--KREF 216
Query 83 PSNSRCIIGLVAGGGRIDKP 102
C+ A GR+ P
Sbjct 217 AFQRTCM----ATVGRLSNP 232
> Hs21265070
Length=305
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 63/135 (46%), Gaps = 12/135 (8%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A E + G I H G+ A A G+ PLG +P G+++ +VE +PG + +G
Sbjct 152 IATENMQAGDTILNSNHIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYIRAAG 211
Query 56 SFATVVGHSEETGKSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLLKAGTAYHKYKA 115
+ ++ + G + ++LPS + + +G V+ + + KAG +++
Sbjct 212 TCGVLL--RKVNGTAIIQLPSKRQMQVLETCVATVGRVSNVDHNKRVIGKAGR--NRWLG 267
Query 116 KR---NCWHKVRGVA 127
KR WH+ G A
Sbjct 268 KRPNSGRWHRKGGWA 282
> HsM7705714
Length=353
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 42/81 (51%), Gaps = 7/81 (8%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A E + G I H G+ A A G+ PLG +P G+++ +VE +PG + +G
Sbjct 152 IATENMQAGDTILNSNHIGRMAVAAREGDAHPLGALPVGTLINNVESEPGRGAQYIRAAG 211
Query 56 SFATVVGHSEETGKSRVRLPS 76
+ ++ + G S ++LPS
Sbjct 212 TCGVLL--RKVNGHSIIQLPS 230
> Hs22061364
Length=197
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 44/81 (54%), Gaps = 7/81 (8%)
Query 1 LAVEGIHTGQFI----HCGKKA-ALSIGNVLPLGKVPEGSVVCSVEEKPGDRGALAKTSG 55
+A++ + G I H G+ A A G+ PLG +P G+++ ++E +PG + +G
Sbjct 57 MALKNLQAGDTILNSNHIGQMAVAAQEGDAHPLGALPVGTLINNMEIEPGRGAQYIQAAG 116
Query 56 SFATVVGHSEETGKSRVRLPS 76
+ + ++ + G + ++LPS
Sbjct 117 TCSVLLW--KVNGTTVIQLPS 135
> 7290363
Length=982
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 3/53 (5%)
Query 1 LAVEGIHTGQFIHCGKKAALSIGNVLPLGKVPEGSV---VCSVEEKPGDRGAL 50
L+V +H+G F + +S + G P+ S+ S +++PGD+GAL
Sbjct 804 LSVRQLHSGLFEPIDRVDGISSAGISTSGSKPDSSMNATTASQDQEPGDQGAL 856
> YGL092w
Length=1317
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 29 GKVPEGSVVCSVEEKPGDRGALAKTSGSFATVVGHSEETGKSRVRLP 75
G+ P V + KP D+ + KT G+F T+ G + + + P
Sbjct 630 GEQPLRKVRTLAQSKPSDKEVILKTDGTFGTLSGKDDSIVEEKAYEP 676
> 7304025_1
Length=326
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query 69 KSRVRLPSGARKTLPSNSRCIIGLVAGGGRIDKPLL 104
K +LP+G T PSN+R + G V G G D LL
Sbjct 246 KPSSKLPAGGNATSPSNTRILTG-VGGSGTDDGTLL 280
Lambda K H
0.316 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40