bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2785_orf1
Length=190
Score E
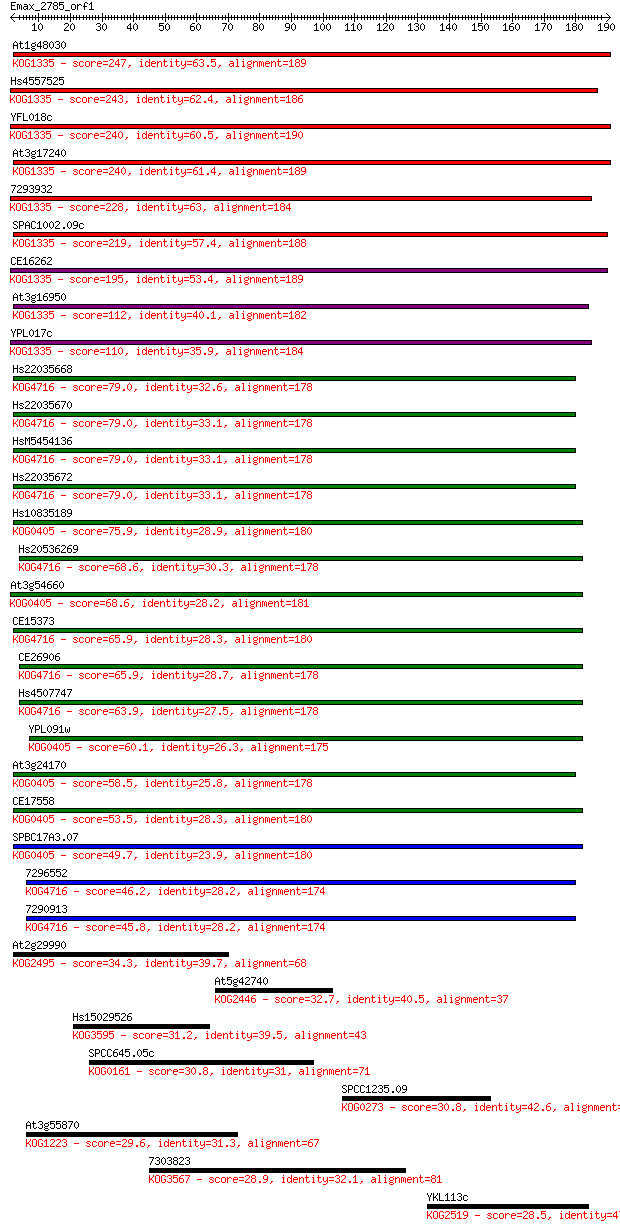
Sequences producing significant alignments: (Bits) Value
At1g48030 247 1e-65
Hs4557525 243 2e-64
YFL018c 240 1e-63
At3g17240 240 1e-63
7293932 228 4e-60
SPAC1002.09c 219 2e-57
CE16262 195 5e-50
At3g16950 112 4e-25
YPL017c 110 2e-24
Hs22035668 79.0 5e-15
Hs22035670 79.0 5e-15
HsM5454136 79.0 5e-15
Hs22035672 79.0 6e-15
Hs10835189 75.9 4e-14
Hs20536269 68.6 8e-12
At3g54660 68.6 8e-12
CE15373 65.9 4e-11
CE26906 65.9 5e-11
Hs4507747 63.9 2e-10
YPL091w 60.1 2e-09
At3g24170 58.5 7e-09
CE17558 53.5 2e-07
SPBC17A3.07 49.7 3e-06
7296552 46.2 4e-05
7290913 45.8 5e-05
At2g29990 34.3 0.17
At5g42740 32.7 0.46
Hs15029526 31.2 1.2
SPCC645.05c 30.8 1.5
SPCC1235.09 30.8 1.9
At3g55870 29.6 3.7
7303823 28.9 6.6
YKL113c 28.5 7.8
> At1g48030
Length=507
Score = 247 bits (630), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 120/189 (63%), Positives = 150/189 (79%), Gaps = 2/189 (1%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P+T L LE++G++ DK GR++VND+ + + AIGD+I GPMLAHKAEE+G+ACVE
Sbjct 316 PFTSGLDLEKIGVETDKAGRILVNDRF-LSNVPGVYAIGDVIPGPMLAHKAEEDGVACVE 374
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATGD 121
+AGK G V +D +P V+YTHPE+A VGKTEEQLK EGVSY+ G FPF ANSRA+A +
Sbjct 375 FIAGK-HGHVDYDKVPGVVYTHPEVASVGKTEEQLKKEGVSYRVGKFPFMANSRAKAIDN 433
Query 122 SDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEAL 181
++GLVK+L KETDKILGV I+ PNAGELI EAVLA+ Y A+SED+ RVCHAHPT+SEAL
Sbjct 434 AEGLVKILADKETDKILGVHIMAPNAGELIHEAVLAINYDASSEDIARVCHAHPTMSEAL 493
Query 182 KEACMGCFD 190
KEA M +D
Sbjct 494 KEAAMATYD 502
> Hs4557525
Length=509
Score = 243 bits (619), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 116/186 (62%), Positives = 148/186 (79%), Gaps = 2/186 (1%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
RP+T++LGLEE+GI+LD RGR+ VN + Q K +I AIGD++ GPMLAHKAE+EGI CV
Sbjct 316 RPFTKNLGLEELGIELDPRGRIPVNTRFQT-KIPNIYAIGDVVAGPMLAHKAEDEGIICV 374
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATG 120
E +AG G + ++ +PSVIYTHPE+A VGK+EEQLK EG+ YK G FPFAANSRA+
Sbjct 375 EGMAG-GAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGIEYKVGKFPFAANSRAKTNA 433
Query 121 DSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEA 180
D+DG+VK+L K TD++LG ILGP AGE++ EA LA+EYGA+ ED+ RVCHAHPTLSEA
Sbjct 434 DTDGMVKILGQKSTDRVLGAHILGPGAGEMVNEAALALEYGASCEDIARVCHAHPTLSEA 493
Query 181 LKEACM 186
+EA +
Sbjct 494 FREANL 499
> YFL018c
Length=499
Score = 240 bits (612), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 115/190 (60%), Positives = 147/190 (77%), Gaps = 2/190 (1%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
RPY LG E++G+++DKRGR+V++D+ K+ HI +GD+ GPMLAHKAEEEGIA V
Sbjct 307 RPYIAGLGAEKIGLEVDKRGRLVIDDQFNS-KFPHIKVVGDVTFGPMLAHKAEEEGIAAV 365
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATG 120
E L G G V ++ IPSV+Y+HPE+A VGKTEEQLK G+ YK G FPFAANSRA+
Sbjct 366 EMLK-TGHGHVNYNNIPSVMYSHPEVAWVGKTEEQLKEAGIDYKIGKFPFAANSRAKTNQ 424
Query 121 DSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEA 180
D++G VK+L +T++ILG I+GPNAGE+IAEA LA+EYGA++ED+ RVCHAHPTLSEA
Sbjct 425 DTEGFVKILIDSKTERILGAHIIGPNAGEMIAEAGLALEYGASAEDVARVCHAHPTLSEA 484
Query 181 LKEACMGCFD 190
KEA M +D
Sbjct 485 FKEANMAAYD 494
> At3g17240
Length=507
Score = 240 bits (612), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 116/189 (61%), Positives = 148/189 (78%), Gaps = 2/189 (1%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P+T L LE++G++ DK GR++VN++ + + AIGD+I GPMLAHKAEE+G+ACVE
Sbjct 316 PFTSGLDLEKIGVETDKGGRILVNERFST-NVSGVYAIGDVIPGPMLAHKAEEDGVACVE 374
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATGD 121
+AGK G V +D +P V+YT+PE+A VGKTEEQLK EGVSY G FPF ANSRA+A
Sbjct 375 FIAGK-HGHVDYDKVPGVVYTYPEVASVGKTEEQLKKEGVSYNVGKFPFMANSRAKAIDT 433
Query 122 SDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEAL 181
++G+VK+L KETDKILGV I+ PNAGELI EAVLA+ Y A+SED+ RVCHAHPT+SEA+
Sbjct 434 AEGMVKILADKETDKILGVHIMSPNAGELIHEAVLAINYDASSEDIARVCHAHPTMSEAI 493
Query 182 KEACMGCFD 190
KEA M +D
Sbjct 494 KEAAMATYD 502
> 7293932
Length=504
Score = 228 bits (582), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 116/184 (63%), Positives = 139/184 (75%), Gaps = 2/184 (1%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
RPYT LGLE +GI D RGR+ VN Q + +I AIGD I GPMLAHKAE+EG+ +
Sbjct 311 RPYTEGLGLEAVGIVKDDRGRIPVNATFQTV-VPNIYAIGDCIHGPMLAHKAEDEGLITI 369
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATG 120
E + G G + ++ +PSV+YTHPE+A VGK+EEQLK EGV+YK G FPF ANSRA+
Sbjct 370 EGING-GHVHIDYNCVPSVVYTHPEVAWVGKSEEQLKQEGVAYKVGKFPFLANSRAKTNN 428
Query 121 DSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEA 180
D+DG VKVL + TDKILG I+GP AGELI EAVLAMEYGAA+ED+ RVCHAHPT SEA
Sbjct 429 DTDGFVKVLADQATDKILGTHIIGPGAGELINEAVLAMEYGAAAEDVARVCHAHPTCSEA 488
Query 181 LKEA 184
L+EA
Sbjct 489 LREA 492
> SPAC1002.09c
Length=511
Score = 219 bits (559), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 108/188 (57%), Positives = 141/188 (75%), Gaps = 2/188 (1%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
PYT LGL+++GI +DK RV+++ + + HI IGD GPMLAHKAE+EGIA VE
Sbjct 319 PYTEGLGLDKLGISMDKSNRVIMDSEYRT-NIPHIRVIGDATLGPMLAHKAEDEGIAAVE 377
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATGD 121
+A KG G V ++ IP+V+YTHPE+A VG TE++ K G+ Y+ G FPF+ANSRA+ D
Sbjct 378 YIA-KGQGHVNYNCIPAVMYTHPEVAWVGITEQKAKESGIKYRIGTFPFSANSRAKTNMD 436
Query 122 SDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEAL 181
+DGLVKV+ ETD++LGV ++GP AGELI EA LA+EYGA++ED+ RVCHAHPTLSEA
Sbjct 437 ADGLVKVIVDAETDRLLGVHMIGPMAGELIGEATLALEYGASAEDVARVCHAHPTLSEAT 496
Query 182 KEACMGCF 189
KEA M +
Sbjct 497 KEAMMAAW 504
> CE16262
Length=464
Score = 195 bits (496), Expect = 5e-50, Method: Compositional matrix adjust.
Identities = 101/189 (53%), Positives = 121/189 (64%), Gaps = 33/189 (17%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
RPYT LGL + IDLD RGRV VN++ Q K I AIGD+IEGPMLAHKAE+EGI CV
Sbjct 301 RPYTEGLGLSNVQIDLDNRGRVPVNERFQT-KVPSIFAIGDVIEGPMLAHKAEDEGILCV 359
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARATG 120
E +AG GV+YK G FPF ANSRA+
Sbjct 360 EGIAG--------------------------------GPGVAYKIGKFPFVANSRAKTNN 387
Query 121 DSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSEA 180
D +G VKVL K+TD++LGV I+GPNAGE+IAEA LAMEYGA++ED+ RVCH HPTLSEA
Sbjct 388 DQEGFVKVLADKQTDRMLGVHIIGPNAGEMIAEATLAMEYGASAEDVARVCHPHPTLSEA 447
Query 181 LKEACMGCF 189
+EA + +
Sbjct 448 FREANLAAY 456
> At3g16950
Length=564
Score = 112 bits (280), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 73/187 (39%), Positives = 105/187 (56%), Gaps = 9/187 (4%)
Query 2 PYTRDLGLEEMG-IDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
P+T LGLE G I +D+R RV+ V ++ IGD MLAH A +GI+ V
Sbjct 360 PFTNGLGLENRGFIPVDERMRVIDGKGTLV---PNLYCIGDANGKLMLAHAASAQGISVV 416
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLK----AEGVSYKRGAFPFAANSRA 116
E+++G+ D + H +IP+ +THPE++ VG TE Q K EG F AN++A
Sbjct 417 EQVSGR-DHVLNHLSIPAACFTHPEISMVGLTEPQAKEKGEKEGFKVSVVKTSFKANTKA 475
Query 117 RATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPT 176
A + +G+ K++ + +ILGV I G +A +LI EA A+ G +D+ HAHPT
Sbjct 476 LAENEGEGIAKMIYRPDNGEILGVHIFGLHAADLIHEASNAIALGTRIQDIKLAVHAHPT 535
Query 177 LSEALKE 183
LSE L E
Sbjct 536 LSEVLDE 542
> YPL017c
Length=499
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 66/191 (34%), Positives = 99/191 (51%), Gaps = 7/191 (3%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
RP + L + +G+D V +LKY HI IGD+ GPMLA KAEE+ I +
Sbjct 292 RPLLKGLDISSIGLDERDFVENVDVQTQSLLKYPHIKPIGDVTLGPMLALKAEEQAIRAI 351
Query 61 ERLAGKG-DGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSRARA- 118
+ + G DGT P+V+Y P++ VG TEE L + Y++G F+ N R
Sbjct 352 QSIGCTGSDGTSNCGFPPNVLYCQPQIGWVGYTEEGLAKARIPYQKGRVLFSQNVRYNTL 411
Query 119 -----TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHA 173
+KVL KILGV ++ +A EL+++A +A+ G + D+ +V
Sbjct 412 LPREENTTVSPFIKVLIDSRDMKILGVHMINDDANELLSQASMAVSLGLTAHDVCKVPFP 471
Query 174 HPTLSEALKEA 184
HP+LSE+ K+A
Sbjct 472 HPSLSESFKQA 482
> Hs22035668
Length=428
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 58/189 (30%), Positives = 90/189 (47%), Gaps = 17/189 (8%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACV 60
P TR L LE+ G+D + ++ D + HI AIGD++EG P L A G V
Sbjct 224 PDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVEGRPELTPIAIMAGRLLV 283
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQ---------LKAEGVSYKRGAFPFA 111
+RL G + +D +P+ ++T E VG +EE+ ++ YK F A
Sbjct 284 QRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPLEFTVA 343
Query 112 ANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGAASEDLGRV 170
++ VK++ +E + +LG+ LGPNAGE+ L ++ GA+ + R
Sbjct 344 GRDASQC------YVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMRT 397
Query 171 CHAHPTLSE 179
HPT SE
Sbjct 398 VGIHPTCSE 406
> Hs22035670
Length=494
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 59/189 (31%), Positives = 89/189 (47%), Gaps = 17/189 (8%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACV 60
P TR L LE+ G+D + ++ D + HI AIGD++EG P L A G V
Sbjct 290 PDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVEGRPELTPIAIMAGRLLV 349
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAE---------GVSYKRGAFPFA 111
+RL G + +D +P+ ++T E VG +EE+ A YK F A
Sbjct 350 QRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPLEFTVA 409
Query 112 ANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGAASEDLGRV 170
++ VK++ +E + +LG+ LGPNAGE+ L ++ GA+ + R
Sbjct 410 GRDASQC------YVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMRT 463
Query 171 CHAHPTLSE 179
HPT SE
Sbjct 464 VGIHPTCSE 472
> HsM5454136
Length=524
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 59/189 (31%), Positives = 89/189 (47%), Gaps = 17/189 (8%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACV 60
P TR L LE+ G+D + ++ D + HI AIGD++EG P L A G V
Sbjct 320 PDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVEGRPELTPIAIMAGRLLV 379
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAE---------GVSYKRGAFPFA 111
+RL G + +D +P+ ++T E VG +EE+ A YK F A
Sbjct 380 QRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPLEFTVA 439
Query 112 ANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGAASEDLGRV 170
++ VK++ +E + +LG+ LGPNAGE+ L ++ GA+ + R
Sbjct 440 GRDASQC------YVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMRT 493
Query 171 CHAHPTLSE 179
HPT SE
Sbjct 494 VGIHPTCSE 502
> Hs22035672
Length=524
Score = 79.0 bits (193), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 59/189 (31%), Positives = 89/189 (47%), Gaps = 17/189 (8%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACV 60
P TR L LE+ G+D + ++ D + HI AIGD++EG P L A G V
Sbjct 320 PDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVEGRPELTPIAIMAGRLLV 379
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAE---------GVSYKRGAFPFA 111
+RL G + +D +P+ ++T E VG +EE+ A YK F A
Sbjct 380 QRLFGGSSDLMDYDNVPTTVFTPLEYGCVGLSEEEAVARHGQEHVEVYHAHYKPLEFTVA 439
Query 112 ANSRARATGDSDGLVKVLTCKETDK-ILGVWILGPNAGELIAEAVLAMEYGAASEDLGRV 170
++ VK++ +E + +LG+ LGPNAGE+ L ++ GA+ + R
Sbjct 440 GRDASQC------YVKMVCLREPPQLVLGLHFLGPNAGEVTQGFALGIKCGASYAQVMRT 493
Query 171 CHAHPTLSE 179
HPT SE
Sbjct 494 VGIHPTCSE 502
> Hs10835189
Length=479
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 52/183 (28%), Positives = 90/183 (49%), Gaps = 4/183 (2%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P T+DL L ++GI D +G ++V D+ Q I A+GD+ +L A G
Sbjct 294 PNTKDLSLNKLGIQTDDKGHIIV-DEFQNTNVKGIYAVGDVCGKALLTPVAIAAGRKLAH 352
Query 62 RL-AGKGDGTVRHDTIPSVIYTHPELAGVGKTE-EQLKAEGV-SYKRGAFPFAANSRARA 118
RL K D + ++ IP+V+++HP + VG TE E + G+ + K + F A
Sbjct 353 RLFEYKEDSKLDYNNIPTVVFSHPPIGTVGLTEDEAIHKYGIENVKTYSTSFTPMYHAVT 412
Query 119 TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLS 178
+ ++K++ + +K++G+ + G E++ +A++ GA D HPT S
Sbjct 413 KRKTKCVMKMVCANKEEKVVGIHMQGLGCDEMLQGFAVAVKMGATKADFDNTVAIHPTSS 472
Query 179 EAL 181
E L
Sbjct 473 EEL 475
> Hs20536269
Length=459
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 54/186 (29%), Positives = 89/186 (47%), Gaps = 11/186 (5%)
Query 4 TRDLGLEEMGIDLD-KRGRVVVNDKMQVLKYAHIMAIGDLIE-GPMLAHKAEEEGIACVE 61
TR +GLE++G+ ++ K G++ VND Q ++ A+GD++E P L A + G +
Sbjct 259 TRKIGLEKIGVKINEKSGKIPVNDVEQT-NVPYVYAVGDILEDKPELTPVAIQSGKLLAQ 317
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQ----LKAEGVS-YKRGAFPFAANSRA 116
RL G + +P+ ++T E G +EE+ K E + Y +P
Sbjct 318 RLFGASLEKCDYINVPTTVFTPLEYGCCGLSEEKAIEVYKKENLEIYHTLFWPLEWTVAG 377
Query 117 RATGDSDGLVKVLTCK-ETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHP 175
R ++ K++ K + D+++G ILGPNAGE+ AM+ G + L HP
Sbjct 378 RE--NNTCYAKIICNKFDHDRVIGFHILGPNAGEVTQGFAAAMKCGLTKQLLDDTIGIHP 435
Query 176 TLSEAL 181
T E
Sbjct 436 TCGEVF 441
> At3g54660
Length=565
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 51/185 (27%), Positives = 87/185 (47%), Gaps = 9/185 (4%)
Query 1 RPYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACV 60
+P T++LGLE +G+ + K G + V++ Q I A+GD+ + L A EG A
Sbjct 357 KPNTKNLGLENVGVKMAKNGAIEVDEYSQT-SVPSIWAVGDVTDRINLTPVALMEGGALA 415
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVSYKRGAFPFAANSR---AR 117
+ L + +P +++ P + VG TEEQ + + +N R A
Sbjct 416 KTLFQNEPTKPDYRAVPCAVFSQPPIGTVGLTEEQ----AIEQYGDVDVYTSNFRPLKAT 471
Query 118 ATGDSDG-LVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPT 176
+G D +K++ C T+K+LGV + G ++ E+I +A++ G D HPT
Sbjct 472 LSGLPDRVFMKLIVCANTNKVLGVHMCGEDSPEIIQGFGVAVKAGLTKADFDATVGVHPT 531
Query 177 LSEAL 181
+E
Sbjct 532 AAEEF 536
> CE15373
Length=503
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 51/186 (27%), Positives = 80/186 (43%), Gaps = 6/186 (3%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPM-LAHKAEEEGIACV 60
P + L L+ G+ DKR ++ D+ + A+GD+++ L A + G
Sbjct 298 PNLKSLNLDNAGVRTDKRSGKILADEFDRASCNGVYAVGDIVQDRQELTPLAIQSGKLLA 357
Query 61 ERLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQ-LKAEGVS----YKRGAFPFAANSR 115
+RL VR D + + ++T EL+ VG TEE+ ++ G + PF
Sbjct 358 DRLFSNSKQIVRFDGVATTVFTPLELSTVGLTEEEAIQKHGEDSIEVFHSHFTPFEYVVP 417
Query 116 ARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHP 175
+ V T E+ KILG+ +GPNA E+I +A G + DL HP
Sbjct 418 QNKDSGFCYVKAVCTRDESQKILGLHFVGPNAAEVIQGYAVAFRVGISMSDLQNTIAIHP 477
Query 176 TLSEAL 181
SE
Sbjct 478 CSSEEF 483
> CE26906
Length=665
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 82/184 (44%), Gaps = 7/184 (3%)
Query 4 TRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACVER 62
T D+GL +G++ K +V+ + Q + AIGD++EG P L A + G + R
Sbjct 464 TDDVGLTTIGVERAKSKKVL-GRREQSTTIPWVYAIGDVLEGTPELTPVAIQAGRVLMRR 522
Query 63 LAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQL-----KAEGVSYKRGAFPFAANSRAR 117
+ + +D IP+ ++T E G +EE K + Y P R
Sbjct 523 IFDGANELTEYDQIPTTVFTPLEYGCCGLSEEDAMMKYGKDNIIIYHNVFNPLEYTISER 582
Query 118 ATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTL 177
D L + E +K++G IL PNAGE+ +A++ A D R+ HPT+
Sbjct 583 MDKDHCYLKMICLRNEEEKVVGFHILTPNAGEVTQGFGIALKLAAKKADFDRLIGIHPTV 642
Query 178 SEAL 181
+E
Sbjct 643 AENF 646
> Hs4507747
Length=497
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 49/187 (26%), Positives = 88/187 (47%), Gaps = 13/187 (6%)
Query 4 TRDLGLEEMGIDLD-KRGRVVVNDKMQVLKYAHIMAIGDLIEGPM-LAHKAEEEGIACVE 61
TR +GLE +G+ ++ K G++ V D+ Q +I AIGD++E + L A + G +
Sbjct 297 TRKIGLETVGVKINEKTGKIPVTDEEQT-NVPYIYAIGDILEDKVELTPVAIQAGRLLAQ 355
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQ----LKAEGVS-YKRGAFPFAANSRA 116
RL ++ +P+ ++T E G +EE+ E + Y +P +
Sbjct 356 RLYAGSTVKCDYENVPTTVFTPLEYGACGLSEEKAVEKFGEENIEVYHSYFWPLEWTIPS 415
Query 117 RATGDSDGLVKVLTC--KETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAH 174
R D++ + C K+ ++++G +LGPNAGE+ A++ G + L H
Sbjct 416 R---DNNKCYAKIICNTKDNERVVGFHVLGPNAGEVTQGFAAALKCGLTKKQLDSTIGIH 472
Query 175 PTLSEAL 181
P +E
Sbjct 473 PVCAEVF 479
> YPL091w
Length=483
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 46/182 (25%), Positives = 85/182 (46%), Gaps = 10/182 (5%)
Query 7 LGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVERLAGK 66
+G E +GI L+ +++ D+ Q +I ++GD++ L A G RL G
Sbjct 301 MGSENVGIKLNSHDQIIA-DEYQNTNVPNIYSLGDVVGKVELTPVAIAAGRKLSNRLFGP 359
Query 67 ---GDGTVRHDTIPSVIYTHPELAGVGKTE----EQLKAEGVSYKRGAFPFAANSRARAT 119
+ + ++ +PSVI++HPE +G +E E+ E + F A A +
Sbjct 360 EKFRNDKLDYENVPSVIFSHPEAGSIGISEKEAIEKYGKENIKVYNSKF--TAMYYAMLS 417
Query 120 GDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLSE 179
S K++ +K++G+ I+G ++ E++ +A++ GA D HPT +E
Sbjct 418 EKSPTRYKIVCAGPNEKVVGLHIVGDSSAEILQGFGVAIKMGATKADFDNCVAIHPTSAE 477
Query 180 AL 181
L
Sbjct 478 EL 479
> At3g24170
Length=499
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 46/182 (25%), Positives = 78/182 (42%), Gaps = 8/182 (4%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P T+ L LE +G++LD+ G V V D+ I A+GD L A E
Sbjct 300 PNTKRLNLEAVGVELDQAGAVKV-DEYSRTNIPSIWAVGDATNRINLTPVALMEATCFAN 358
Query 62 RLAGKGDGTVRHDTIPSVIYTHPELAGVGKTEE----QLKAEGVSYKRGAFPFAANSRAR 117
G + + ++ P LA VG +EE Q + + + G P R
Sbjct 359 TAFGGKPTKAEYSNVACAVFCIPPLAVVGLSEEEAVEQATGDILVFTSGFNPMKNTISGR 418
Query 118 ATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTL 177
L+K++ +++DK++G + GP+A E++ +A++ GA HP+
Sbjct 419 ---QEKTLMKLIVDEKSDKVIGASMCGPDAAEIMQGIAIALKCGATKAQFDSTVGIHPSS 475
Query 178 SE 179
+E
Sbjct 476 AE 477
> CE17558
Length=473
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 51/188 (27%), Positives = 88/188 (46%), Gaps = 14/188 (7%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P T++L LE +G+ DK G ++V D+ Q I+++GD +L A G
Sbjct 282 PLTKELNLERVGVKTDKSGHIIV-DEYQNTSAPGILSVGDDTGKFLLTPVAIAAGRRLSH 340
Query 62 RL-AGKGDGTVRHDTIPSVIYTHPELAGVGKTEEQL-----KAEGVSYKRGAFP--FAAN 113
RL G+ D + ++ I +V+++HP + VG TE + K E YK P FA
Sbjct 341 RLFNGETDNKLTYENIATVVFSHPLIGTVGLTEAEAVEKYGKDEVTLYKSRFNPMLFAVT 400
Query 114 SRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHA 173
+K++ + +K++GV + G + E++ +A+ GA + +
Sbjct 401 KHKEKAA-----MKLVCVGKDEKVVGVHVFGVGSDEMLQGFAVAVTMGATKKQFDQTVAI 455
Query 174 HPTLSEAL 181
HPT +E L
Sbjct 456 HPTSAEEL 463
> SPBC17A3.07
Length=464
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/190 (22%), Positives = 80/190 (42%), Gaps = 16/190 (8%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVE 61
P + L LE+ G+ G +++ D Q ++++GD+ L A G +
Sbjct 277 PKIQGLRLEKAGVKTLPNG-IIIADTYQRTNVPTVLSLGDVCGKLELTPVAIAAGRRLSD 335
Query 62 RL-AGKGDGTVRHDTIPSVIYTHPELAGVGKTEE---------QLKAEGVSYKRGAFPFA 111
RL G D + ++ +PSV++ HPE +G TE+ Q+K + +
Sbjct 336 RLFGGIKDAHLDYEEVPSVVFAHPEAGTIGLTEQEAIDKYGESQIKVYNTKFNGLNYSMV 395
Query 112 ANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVC 171
T K++ K++G+ ++G + E++ +A++ GA D
Sbjct 396 EQEDKVPT-----TYKLVCAGPLQKVVGLHLVGDFSAEILQGFGVAIKMGATKSDFDSCV 450
Query 172 HAHPTLSEAL 181
HPT +E L
Sbjct 451 AIHPTSAEEL 460
> 7296552
Length=516
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 49/181 (27%), Positives = 77/181 (42%), Gaps = 11/181 (6%)
Query 6 DLGLEEMGIDL--DKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACVER 62
DL L+ G+ DK +V D + HI A+GD+I G P L A G R
Sbjct 318 DLNLDAAGVKTHDDK----IVVDAAEATSVPHIFAVGDIIYGRPELTPVAILSGRLLARR 373
Query 63 LAGKGDGTVRHDTIPSVIYTHPELAGVGKTEE---QLK-AEGVSYKRGAFPFAANSRARA 118
L + + + + ++T E + VG +EE +L+ A+ + G + +
Sbjct 374 LFAGSTQLMDYADVATTVFTPLEYSCVGMSEETAIELRGADNIEVFHGYYKPTEFFIPQK 433
Query 119 TGDSDGLVKVLTCKETDKILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHPTLS 178
+ L V KILG+ +GP AGE+I A++ G + L HPT +
Sbjct 434 SVRHCYLKAVAEVSGDQKILGLHYIGPVAGEVIQGFAAALKTGLTVKTLLNTVGIHPTTA 493
Query 179 E 179
E
Sbjct 494 E 494
> 7290913
Length=491
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 49/184 (26%), Positives = 76/184 (41%), Gaps = 17/184 (9%)
Query 6 DLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEG-PMLAHKAEEEGIACVERLA 64
DL L G+ + K V D + A+I A+GD+I G P L A G RL
Sbjct 293 DLNLPNAGVTVQKDKIPV--DSQEATNVANIYAVGDIIYGKPELTPVAVLAGRLLARRLY 350
Query 65 GKGDGTVRHDTIPSVIYTHPELAGVGKTEE----QLKAEGVSYKRGAFP----FAANSRA 116
G + + + + ++T E A VG +EE Q A+ + G + F
Sbjct 351 GGSTQRMDYKDVATTVFTPLEYACVGLSEEDAVKQFGADEIEVFHGYYKPTEFFIPQKSV 410
Query 117 RATGDSDGLVKVLTCKETD-KILGVWILGPNAGELIAEAVLAMEYGAASEDLGRVCHAHP 175
R +K + + D ++ G+ +GP AGE+I A++ G L HP
Sbjct 411 RYC-----YLKAVAERHGDQRVYGLHYIGPVAGEVIQGFAAALKSGLTINTLINTVGIHP 465
Query 176 TLSE 179
T +E
Sbjct 466 TTAE 469
> At2g29990
Length=508
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 16/79 (20%)
Query 2 PYTRDLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGD---LIEG------PMLAHKA 52
P+ R LGL + D GR+ +++ M+V + AIGD +E P LA A
Sbjct 346 PFVRSLGLPK-----DPTGRIGIDEWMRVPSVQDVFAIGDCSGYLETTGKPTLPALAQVA 400
Query 53 EEEG--IACVERLAGKGDG 69
E EG +A + GKG+G
Sbjct 401 EREGKYLANLLNAIGKGNG 419
> At5g42740
Length=560
Score = 32.7 bits (73), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 66 KGDGTVRHDTIPSVIYTHPELAGVGKTEEQLKAEGVS 102
KG+ HD + S + P+ GKT EQL+ E VS
Sbjct 421 KGEVVSNHDELMSNFFAQPDALAYGKTPEQLQKENVS 457
> Hs15029526
Length=4490
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 2/43 (4%)
Query 21 RVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVERL 63
++ +N KMQ+L+ +I+ +L+EG L EE G++CVE L
Sbjct 2306 KLNLNPKMQLLECNYIVQSLNLLEG--LIPSKEEGGVSCVEHL 2346
> SPCC645.05c
Length=1526
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Query 26 DKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVERLAGKGDGTVRHDTIPSVIYTHPE 85
DK Q K H+ AI DL +L +K + + E AGK + T R + I +
Sbjct 136 DKTQERKLPHVFAIADLAYNNLLENKENQSILVTGESGAGKTENTKRIIQYLAAIASSTT 195
Query 86 LAGVGKTEEQL 96
+ G + EEQ+
Sbjct 196 V-GSSQVEEQI 205
> SPCC1235.09
Length=564
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 106 GAFPFAANSRARATGDSDGLVKVLTCKETDKILGVWILGPNAGELIA 152
A F+ N R ATGD+ G V + +CK T K+ LG + ELIA
Sbjct 479 SALSFSHNGRYLATGDTSGGVCIWSCK-TAKLFKE--LGSDNSELIA 522
> At3g55870
Length=526
Score = 29.6 bits (65), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 8/68 (11%)
Query 6 DLGLEEMGIDLDKRGRVVVNDKMQVLKYAHIMAIGDLIEGPMLAHKAEEEGIACVERL-A 64
DLG ++G + K G V V M + +Y+H+M I + G + +E + C + L A
Sbjct 355 DLGRNDVG-KVSKNGSVKVERLMNIERYSHVMHISSTVIGEL------QENLTCWDTLRA 407
Query 65 GKGDGTVR 72
GTV+
Sbjct 408 ALPVGTVK 415
> 7303823
Length=541
Score = 28.9 bits (63), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 33/98 (33%), Gaps = 17/98 (17%)
Query 45 GPMLAHKAEEEGIACVERLAGKGDG-------TVRHDTIPSVIYTHPELAGVGKTEEQLK 97
PM HK +A L+ D V D +P+V HP + L
Sbjct 408 NPMRIHKFVHRSLAKPMSLSASKDSRDSAISQAVGGDQVPAVAVHHPSGKAILVASLMLL 467
Query 98 AEGVSY----------KRGAFPFAANSRARATGDSDGL 125
G ++ KRG PF A R A SDG
Sbjct 468 FAGSTFALALIFARRRKRGCLPFGARGRRHAWEKSDGF 505
> YKL113c
Length=382
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 32/64 (50%), Gaps = 14/64 (21%)
Query 133 ETDKILGV----WILGPNAGELIAEAVLAME---YGAASEDLGRVCHAHP------TLSE 179
E K+LG+ +I+ P E A LA + Y AASED+ +C+ P T SE
Sbjct 138 EAQKLLGLMGIPYIIAPTEAEAQC-AELAKKGKVYAAASEDMDTLCYRTPFLLRHLTFSE 196
Query 180 ALKE 183
A KE
Sbjct 197 AKKE 200
Lambda K H
0.318 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3166472848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40