bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2836_orf1
Length=56
Score E
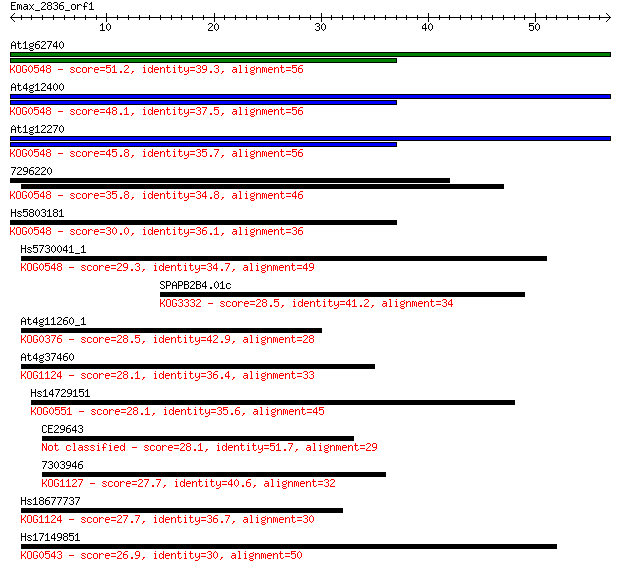
Sequences producing significant alignments: (Bits) Value
At1g62740 51.2 4e-07
At4g12400 48.1 4e-06
At1g12270 45.8 2e-05
7296220 35.8 0.019
Hs5803181 30.0 1.1
Hs5730041_1 29.3 2.0
SPAPB2B4.01c 28.5 3.1
At4g11260_1 28.5 3.3
At4g37460 28.1 4.0
Hs14729151 28.1 4.5
CE29643 28.1 4.7
7303946 27.7 5.1
Hs18677737 27.7 6.2
Hs17149851 26.9 8.5
> At1g62740
Length=571
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVVRKVQQQQSGEIDEEQ 56
SRKG++ F +KEY A+E + KGL DPN++E +DG + V+++ + G++ E+
Sbjct 454 SRKGAVQFFMKEYDNAMETYQKGLEHDPNNQELLDGVKRCVQQINKANRGDLTPEE 509
Score = 34.7 bits (78), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDG 36
SR G+ H L ++ +A+EA+ KGL IDP+++ G
Sbjct 74 SRLGAAHLGLNQFDEAVEAYSKGLEIDPSNEGLKSG 109
> At4g12400
Length=558
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVVRKVQQQQSGEIDEEQ 56
SRKG++ F +KEY KA+E + +GL DP ++E +DG + V ++ + G++ E+
Sbjct 441 SRKGAIQFFMKEYDKAMETYQEGLKHDPKNQEFLDGVRRCVEQINKASRGDLTPEE 496
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDG 36
SR G+ L ++ +A++++ KGL IDP+++ G
Sbjct 74 SRLGAAFIGLSKFDEAVDSYKKGLEIDPSNEMLKSG 109
> At1g12270
Length=572
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 36/56 (64%), Gaps = 0/56 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVVRKVQQQQSGEIDEEQ 56
SRK ++ F LKEY A+E + GL DP+++E +DG + V+++ + G++ E+
Sbjct 455 SRKAAVQFFLKEYDNAMETYQAGLEHDPSNQELLDGVKRCVQQINKANRGDLTPEE 510
Score = 31.2 bits (69), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDG 36
SR G+ H L ++ A+ A+ KGL +DP ++ G
Sbjct 74 SRLGAAHLGLNQFELAVTAYKKGLDVDPTNEALKSG 109
> 7296220
Length=490
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVV 41
SRKG+ L ++ KA EA+++GL DP + + G+M+
Sbjct 76 SRKGAAAAGLNDFMKAFEAYNEGLKYDPTNAILLQGRMETT 116
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVVRKVQQ 46
RKG + +++ +KA A+ K L +DPN+ E ++G Q Q+
Sbjct 383 RKGKILQGMQQQSKAQAAYQKALELDPNNAEAIEGYRQCSMNFQR 427
> Hs5803181
Length=543
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 1 SRKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDG 36
+RK + +K+Y KA++ + K L +D + KE DG
Sbjct 432 TRKAAALEAMKDYTKAMDVYQKALDLDSSCKEAADG 467
> Hs5730041_1
Length=137
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQV-VRKVQQQQSG 50
RKG + K YA ALE F +G +D D V +++ Q+ Q+G
Sbjct 84 RKGICEYHEKNYAAALETFTEGQKLDS-----ADANFSVWIKRCQEAQNG 128
> SPAPB2B4.01c
Length=248
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query 15 KALEAFD-KGLAIDPNSKECMDGKMQVVRKVQQQQ 48
K L FD KG++ PN C +G M++V+ Q Q
Sbjct 133 KTLITFDNKGISGHPNHIACYEGAMKIVKATPQVQ 167
> At4g11260_1
Length=160
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPN 29
RKG+ +L+EY+ A A +KG ++ PN
Sbjct 75 RKGTACMKLEEYSTAKAALEKGASVAPN 102
> At4g37460
Length=1013
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPNSKECM 34
R+G L EY +A+E K L +PNS + +
Sbjct 370 RRGQARAALGEYVEAVEDLTKALVFEPNSPDVL 402
> Hs14729151
Length=387
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Query 3 KGSL-HFQLKEYAKALEAFDKGLAIDPNSKEC--MDGKMQVVRKVQQQ 47
+G+L H +LK +A+A+ D+GL ID K+ M K +++++Q+
Sbjct 156 RGALCHLELKHFAEAVNWCDEGLQIDAKEKKLLEMRAKADKLKRIEQR 203
> CE29643
Length=603
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 1/30 (3%)
Query 4 GSLHFQLKEYAKALEAFDKGL-AIDPNSKE 32
G+L F+ K+Y ALEAF KG+ PNS +
Sbjct 51 GNLKFKEKQYDSALEAFTKGVEKAGPNSSD 80
> 7303946
Length=1119
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 16/32 (50%), Gaps = 0/32 (0%)
Query 4 GSLHFQLKEYAKALEAFDKGLAIDPNSKECMD 35
G LH LK Y+ AL K + P+ EC D
Sbjct 413 GKLHMALKNYSDALNYVLKATRLRPHFAECFD 444
> Hs18677737
Length=420
Score = 27.7 bits (60), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPNSK 31
R+G+ QL+ Y + L+ ++ L IDP++K
Sbjct 371 RRGTAFCQLELYVEGLQDYEAALKIDPSNK 400
> Hs17149851
Length=356
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 2 RKGSLHFQLKEYAKALEAFDKGLAIDPNSKECMDGKMQVVRKVQQQQSGE 51
RKG + Q EY++A+ L ++P++K ++V+K Q+S E
Sbjct 255 RKGKVLAQQGEYSEAIPILRAALKLEPSNKTIHAELSKLVKKHAAQRSTE 304
Lambda K H
0.311 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194096762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40