bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2860_orf2
Length=56
Score E
Sequences producing significant alignments: (Bits) Value
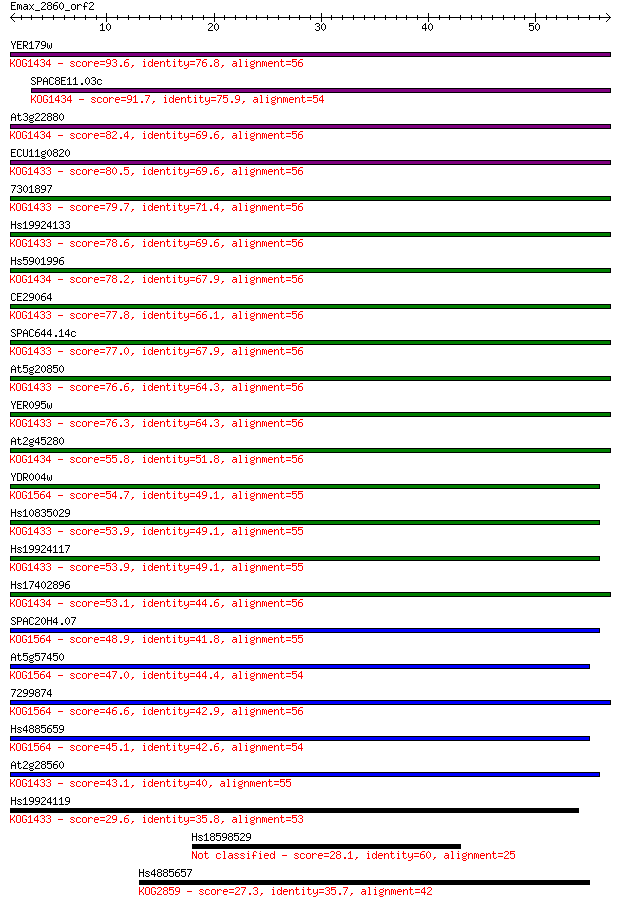
YER179w 93.6 9e-20
SPAC8E11.03c 91.7 3e-19
At3g22880 82.4 2e-16
ECU11g0820 80.5 7e-16
7301897 79.7 1e-15
Hs19924133 78.6 2e-15
Hs5901996 78.2 4e-15
CE29064 77.8 4e-15
SPAC644.14c 77.0 7e-15
At5g20850 76.6 1e-14
YER095w 76.3 1e-14
At2g45280 55.8 2e-08
YDR004w 54.7 4e-08
Hs10835029 53.9 8e-08
Hs19924117 53.9 8e-08
Hs17402896 53.1 1e-07
SPAC20H4.07 48.9 3e-06
At5g57450 47.0 9e-06
7299874 46.6 1e-05
Hs4885659 45.1 4e-05
At2g28560 43.1 1e-04
Hs19924119 29.6 1.3
Hs18598529 28.1 4.2
Hs4885657 27.3 6.4
> YER179w
Length=334
Score = 93.6 bits (231), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 43/56 (76%), Positives = 47/56 (83%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D +LGGGI +MSI E+FGE RCGKTQ+ HTLCVT QLPREMGGG GKV YIDTE T
Sbjct 104 DSILGGGIMTMSITEVFGEFRCGKTQMSHTLCVTTQLPREMGGGEGKVAYIDTEGT 159
> SPAC8E11.03c
Length=332
Score = 91.7 bits (226), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 41/54 (75%), Positives = 47/54 (87%), Gaps = 0/54 (0%)
Query 3 LLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
+LGGGI+SMSI E+FGE RCGKTQ+ HTLCVTAQLPR+MGG GKV +IDTE T
Sbjct 105 ILGGGIQSMSITEVFGEFRCGKTQMSHTLCVTAQLPRDMGGAEGKVAFIDTEGT 158
> At3g22880
Length=339
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/56 (69%), Positives = 42/56 (75%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D LLGGGIE+ +I E FGE R GKTQ+ HTLCVT QLP M GG GKV YIDTE T
Sbjct 116 DDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVTTQLPTNMKGGNGKVAYIDTEGT 171
> ECU11g0820
Length=334
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 39/56 (69%), Positives = 42/56 (75%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+LL GG ES SI EIFGE R GKTQ+CHT+ VT QLP E GGG GK YIDTE T
Sbjct 104 DKLLSGGFESGSITEIFGEFRTGKTQLCHTVAVTCQLPPEQGGGGGKAMYIDTEGT 159
> 7301897
Length=336
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/56 (71%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+LLGGGIE+ SI EIFGE RCGKTQ+CHTL VT QLP GG GK YIDTE T
Sbjct 107 DKLLGGGIETGSITEIFGEFRCGKTQLCHTLAVTCQLPISQKGGEGKCMYIDTENT 162
> Hs19924133
Length=339
Score = 78.6 bits (192), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 39/56 (69%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+LL GGIE+ SI E+FGE R GKTQICHTL VT QLP + GGG GK YIDTE T
Sbjct 110 DKLLQGGIETGSITEMFGEFRTGKTQICHTLAVTCQLPIDRGGGEGKAMYIDTEGT 165
> Hs5901996
Length=340
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/56 (67%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+LLGGGIESM+I E FGE R GKTQ+ HTLCVTAQLP G GK+ +IDTE T
Sbjct 109 DKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPGAGGYPGGKIIFIDTENT 164
> CE29064
Length=395
Score = 77.8 bits (190), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 37/56 (66%), Positives = 44/56 (78%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+LLGGGIE+ SI E++GE R GKTQ+CH+L V QLP +MGGG GK YIDT AT
Sbjct 164 DRLLGGGIETGSITEVYGEYRTGKTQLCHSLAVLCQLPIDMGGGEGKCMYIDTNAT 219
> SPAC644.14c
Length=365
Score = 77.0 bits (188), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 38/56 (67%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D LL GG+E+ SI E+FGE R GK+QICHTL VT QLP +MGGG GK YIDTE T
Sbjct 132 DTLLQGGVETGSITELFGEFRTGKSQICHTLAVTCQLPIDMGGGEGKCLYIDTEGT 187
> At5g20850
Length=339
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 43/56 (76%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D++L GGIE+ SI E++GE R GKTQ+CHTLCVT QLP + GGG GK YID E T
Sbjct 110 DKVLEGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPMDQGGGEGKAMYIDAEGT 165
> YER095w
Length=400
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 44/56 (78%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D LLGGG+E+ SI E+FGE R GK+Q+CHTL VT Q+P ++GGG GK YIDTE T
Sbjct 168 DTLLGGGVETGSITELFGEFRTGKSQLCHTLAVTCQIPLDIGGGEGKCLYIDTEGT 223
> At2g45280
Length=332
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/56 (51%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D +LGGGI + EI G GKTQI L V Q+PRE GG GK YIDTE +
Sbjct 83 DNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIPRECGGLGGKAIYIDTEGS 138
> YDR004w
Length=460
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 55
D+LLGGGI + I EIFGE+ GK+Q+ L ++ QL GG GK YI TE
Sbjct 108 DELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYITTEG 162
> Hs10835029
Length=350
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 55
D+ L GG+ S+ EI G CGKTQ C + + A LP MGG G V YIDTE+
Sbjct 91 DEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTES 145
> Hs19924117
Length=350
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 55
D+ L GG+ S+ EI G CGKTQ C + + A LP MGG G V YIDTE+
Sbjct 91 DEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTES 145
> Hs17402896
Length=376
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D +LGGG+ M EI G GKTQ+C L V Q+P GG G+ +IDTE +
Sbjct 108 DDILGGGVPLMKTTEICGAPGVGKTQLCMQLAVDVQIPECFGGVAGEAVFIDTEGS 163
> SPAC20H4.07
Length=354
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/55 (41%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 55
D+ L GGI + EI GE+ GK+Q C LC+ QLP +GG +I TE+
Sbjct 83 DETLHGGIPVGQLTEICGESGSGKSQFCMQLCLMVQLPLSLGGMNKAAVFISTES 137
> At5g57450
Length=304
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 24/54 (44%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTE 54
D L GGI S+ EI E+ CGKTQ+C L + QLP GG G Y+ +E
Sbjct 29 DGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYLHSE 82
> 7299874
Length=341
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 33/56 (58%), Gaps = 0/56 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 56
D+ GGG+ + I E+ G GKTQ+ L + QLPRE+GG V YI TE++
Sbjct 96 DRCTGGGVVTRGITELCGAAGVGKTQLLLQLSLCVQLPRELGGLGKGVAYICTESS 151
> Hs4885659
Length=346
Score = 45.1 bits (105), Expect = 4e-05, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTE 54
D LL GG+ I E+ G + GKTQ+ LC+ Q PR+ GG YI TE
Sbjct 90 DALLRGGLPLDGITELAGRSSAGKTQLALQLCLAVQFPRQHGGLEAGAVYICTE 143
> At2g28560
Length=353
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 55
D L GGI + E+ G GK+Q C L ++A P GG G+V YID E+
Sbjct 93 DDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVES 147
> Hs19924119
Length=328
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 1 DQLLGGGIESMSIAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDT 53
D+LL G+ + + EI G GKTQ+C LC+ A + G V Y+D+
Sbjct 90 DKLLDAGLYTGEVTEIVGGPGSGKTQVC--LCMAANVAH---GLQQNVLYVDS 137
> Hs18598529
Length=116
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Query 18 GENRCGKTQICHTLCVTAQL-PREMG 42
GE RC K QICH L + L P EMG
Sbjct 72 GEERCPKRQICHKLGLLIMLHPTEMG 97
> Hs4885657
Length=280
Score = 27.3 bits (59), Expect = 6.4, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 13 IAEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTE 54
I E G GKT++ + L LP+ GG +V +IDT+
Sbjct 43 ILEFHGPEGTGKTEMLYHLTARCILPKSEGGLEVEVLFIDTD 84
Lambda K H
0.321 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194096762
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40