bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2876_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
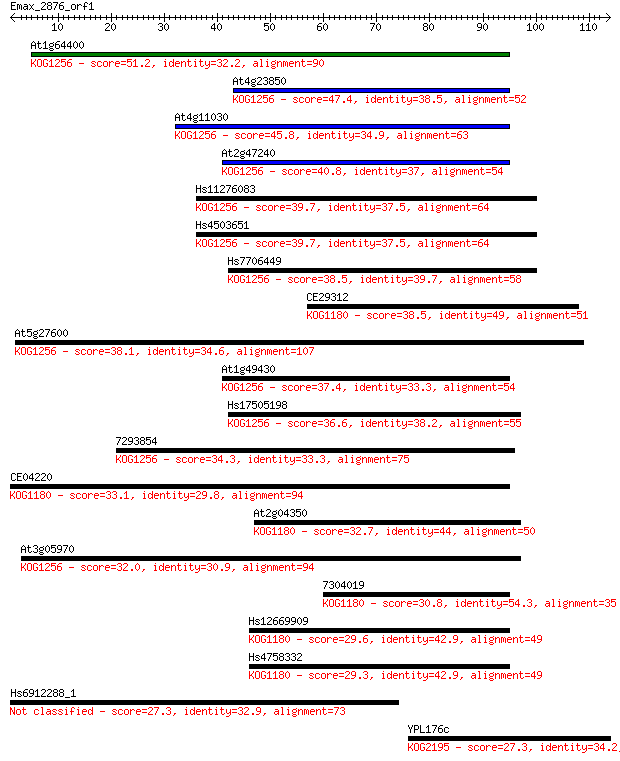
At1g64400 51.2 5e-07
At4g23850 47.4 7e-06
At4g11030 45.8 2e-05
At2g47240 40.8 6e-04
Hs11276083 39.7 0.001
Hs4503651 39.7 0.001
Hs7706449 38.5 0.003
CE29312 38.5 0.003
At5g27600 38.1 0.004
At1g49430 37.4 0.006
Hs17505198 36.6 0.012
7293854 34.3 0.055
CE04220 33.1 0.13
At2g04350 32.7 0.17
At3g05970 32.0 0.29
7304019 30.8 0.57
Hs12669909 29.6 1.4
Hs4758332 29.3 1.7
Hs6912288_1 27.3 6.8
YPL176c 27.3 6.9
> At1g64400
Length=665
Score = 51.2 bits (121), Expect = 5e-07, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 50/95 (52%), Gaps = 8/95 (8%)
Query 5 QYSVSVPSGESTADRTPPR-----RHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGP 59
+Y V V G+ D P R + D E + + +AW++F++ V KS P
Sbjct 5 RYIVEVEKGKQGVDGGSPSVGPVYRSIYAKDGFPE--PPDDLVSAWDIFRLSVEKSPNNP 62
Query 60 CLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
LG R + DG+ G+YVW++Y+EV + ++ G+ +
Sbjct 63 MLGRREIV-DGKAGKYVWQTYKEVHNVVIKLGNSI 96
> At4g23850
Length=666
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 34/52 (65%), Gaps = 1/52 (1%)
Query 43 NAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ W++F+M V K P LG R + DG+ G+YVW++Y+EV + ++ G+ L
Sbjct 46 SCWDVFRMSVEKYPNNPMLGRREIV-DGKPGKYVWQTYQEVYDIVMKLGNSL 96
> At4g11030
Length=666
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 40/66 (60%), Gaps = 4/66 (6%)
Query 32 LAENFSTNPV---KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLAL 88
A+N NP+ ++ W++F+ V K LG R ++ +G+ G+YVWK+Y+EV + +
Sbjct 32 FAQNGFPNPIDGIQSCWDIFRTAVEKYPNNRMLGRREIS-NGKAGKYVWKTYKEVYDIVI 90
Query 89 EAGSGL 94
+ G+ L
Sbjct 91 KLGNSL 96
> At2g47240
Length=660
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ AW++F V K + LG R + D + G Y+WK+Y+EV L+ GS L
Sbjct 41 ITTAWDIFSKSVEKFPDNNMLGWRRIV-DEKVGPYMWKTYKEVYEEVLQIGSAL 93
> Hs11276083
Length=698
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 5/64 (7%)
Query 36 FSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLL 95
+ + V +E FQ G+ S GPCLG+R PD Y W SY++V L+ GS L+
Sbjct 84 YFYDDVTTLYEGFQRGIQVSNNGPCLGSR--KPD---QPYEWLSYKQVAELSECIGSALI 138
Query 96 NGDF 99
F
Sbjct 139 QKGF 142
> Hs4503651
Length=699
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 5/64 (7%)
Query 36 FSTNPVKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLL 95
+ + V +E FQ G+ S GPCLG+R PD Y W SY++V L+ GS L+
Sbjct 84 YFYDDVTTLYEGFQRGIQVSNNGPCLGSR--KPD---QPYEWLSYKQVAELSECIGSALI 138
Query 96 NGDF 99
F
Sbjct 139 QKGF 142
> Hs7706449
Length=683
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 31/58 (53%), Gaps = 5/58 (8%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLNGDF 99
K +E+FQ G+ S GPCLG R P+ Y W SY++V A GS LL+ +
Sbjct 75 KTMYEVFQRGLAVSDNGPCLGYR--KPN---QPYRWLSYKQVSDRAEYLGSCLLHKGY 127
> CE29312
Length=731
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 31/64 (48%), Gaps = 18/64 (28%)
Query 57 EGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGSGLLNGDFIPLL 103
E PCLGTR + P GR GEYVW+SY+EVE +G+ + L
Sbjct 108 ESPCLGTRQLIEVHEDKQPGGRVFEKWHLGEYVWQSYKEVETQVARLAAGIKD-----LA 162
Query 104 HFEQ 107
H EQ
Sbjct 163 HEEQ 166
> At5g27600
Length=698
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 46/113 (40%), Gaps = 8/113 (7%)
Query 2 KDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNP-VKNAWELFQMGVGKSAEGPC 60
K+ YSV +P T R S +L F +P + + F V AE
Sbjct 44 KEDSYSVVLPEKLDTGKWNVYRSK-RSPTKLVSRFPDHPEIGTLHDNFVHAVETYAENKY 102
Query 61 LGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLL-----NGDFIPLLHFEQP 108
LGTR V DG GEY W +Y E GSGLL GD + L +P
Sbjct 103 LGTR-VRSDGTIGEYSWMTYGEAASERQAIGSGLLFHGVNQGDCVGLYFINRP 154
> At1g49430
Length=666
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 41 VKNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGL 94
+ + W+ F V K LG R VT D + G Y W +Y+E A+ GS +
Sbjct 44 IDSPWQFFSEAVKKYPNEQMLGQR-VTTDSKVGPYTWITYKEAHDAAIRIGSAI 96
> Hs17505198
Length=697
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 5/55 (9%)
Query 42 KNAWELFQMGVGKSAEGPCLGTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
+ +++F+ G+ S GPCLG R + Y W SY+EV A GSGLL
Sbjct 90 RTMYQVFRRGLSISGNGPCLGFR-----KPKQPYQWLSYQEVADRAEFLGSGLLQ 139
> 7293854
Length=704
Score = 34.3 bits (77), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 13/82 (15%)
Query 21 PPRRHCSSLDRLAEN-----FSTNPVKNAWELFQMGVGKSAEGPCLGTR--LVTPDGRRG 73
P + H S + ++N + T V+ ++ F+ G S GPCLG R L +P
Sbjct 76 PEQIHVSKFYKESKNGKFVSYITENVRTLYQTFREGAYASNNGPCLGWRETLTSP----- 130
Query 74 EYVWKSYREVERLALEAGSGLL 95
Y W +Y E A G+G+L
Sbjct 131 -YQWINYDEALLRAKNFGAGML 151
> CE04220
Length=726
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 14/107 (13%)
Query 1 AKDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWELFQMGVGKSAEGPC 60
A+ ++ S V + D T P ++ S+ D L N S + + ++ V + PC
Sbjct 49 AEKLEKSERVKAEPDNGDPTTPWKNVSAKDGLM-NSSFEGIHDLGTQWEECVRRYGNQPC 107
Query 61 LGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGSGL 94
+GTR V +G+ G+Y W+SY +V++ SGL
Sbjct 108 MGTREVLKVHKEKQSNGKIFEKWQMGDYHWRSYAQVDKRVNMIASGL 154
> At2g04350
Length=720
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 29/63 (46%), Gaps = 13/63 (20%)
Query 47 LFQMGVGKSAEGPCLGTR-------LVTPDGRR------GEYVWKSYREVERLALEAGSG 93
LF+ K ++ LGTR + DGR+ GEY W+SY EV SG
Sbjct 86 LFEQSCKKYSKDRLLGTREFIDKEFITASDGRKFEKLHLGEYKWQSYGEVFERVCNFASG 145
Query 94 LLN 96
L+N
Sbjct 146 LVN 148
> At3g05970
Length=657
Score = 32.0 bits (71), Expect = 0.29, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 41/95 (43%), Gaps = 3/95 (3%)
Query 3 DVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNP-VKNAWELFQMGVGKSAEGPCL 61
D YSV +P +T R S +L F +P + + F+ V + L
Sbjct 45 DNGYSVVLPEKLNTGSWNVYRSAKSPF-KLVSRFPDHPDIATLHDNFEHAVHDFRDYKYL 103
Query 62 GTRLVTPDGRRGEYVWKSYREVERLALEAGSGLLN 96
GTR V DG G+Y W +Y E GSGL++
Sbjct 104 GTR-VRVDGTVGDYKWMTYGEAGTARTALGSGLVH 137
> 7304019
Length=766
Score = 30.8 bits (68), Expect = 0.57, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 13/48 (27%)
Query 60 CLGTRL-------VTPDGR------RGEYVWKSYREVERLALEAGSGL 94
CLGTR V +GR G+Y WK++ E ER A G GL
Sbjct 103 CLGTRQILSEEDEVQQNGRVFKKYNLGDYKWKTFTEAERTAANFGRGL 150
> Hs12669909
Length=711
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 27/62 (43%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGS 92
+LF V K + LGTR + P+G+ G Y W +Y EV R GS
Sbjct 81 KLFDHAVSKFGKKDSLGTREILSEENEMQPNGKVFKKLILGNYKWMNYLEVNRRVNNFGS 140
Query 93 GL 94
GL
Sbjct 141 GL 142
> Hs4758332
Length=670
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 27/62 (43%), Gaps = 13/62 (20%)
Query 46 ELFQMGVGKSAEGPCLGTRLV-------TPDGR------RGEYVWKSYREVERLALEAGS 92
+LF V K + LGTR + P+G+ G Y W +Y EV R GS
Sbjct 40 KLFDHAVSKFGKKDSLGTREILSEENEMQPNGKVFKKLILGNYKWMNYLEVNRRVNNFGS 99
Query 93 GL 94
GL
Sbjct 100 GL 101
> Hs6912288_1
Length=1923
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 31/75 (41%), Gaps = 16/75 (21%)
Query 1 AKDVQYSVSVPSGESTADRTPPRRHCSSLDRLAENFSTNPVKNAWEL--FQMGVGKSAEG 58
+KDV +S S+P+ E +R S L E TN V W +Q+G G S E
Sbjct 186 SKDVHHSTSLPNLEKEGKPHSDKRSTSHLPTSVEKHCTNGV---WSRSHYQVGEGSSNE- 241
Query 59 PCLGTRLVTPDGRRG 73
D RRG
Sbjct 242 ----------DSRRG 246
> YPL176c
Length=783
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 76 VWKSYREVERLALEAGSGLLNGDFIPLLHFEQPGGQQL 113
++ + ++ +A GLLNGDFI L+H+ QQ+
Sbjct 251 IYANKASLDDMASMQDQGLLNGDFILLVHYGDYVFQQM 288
Lambda K H
0.315 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1185472426
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40