bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
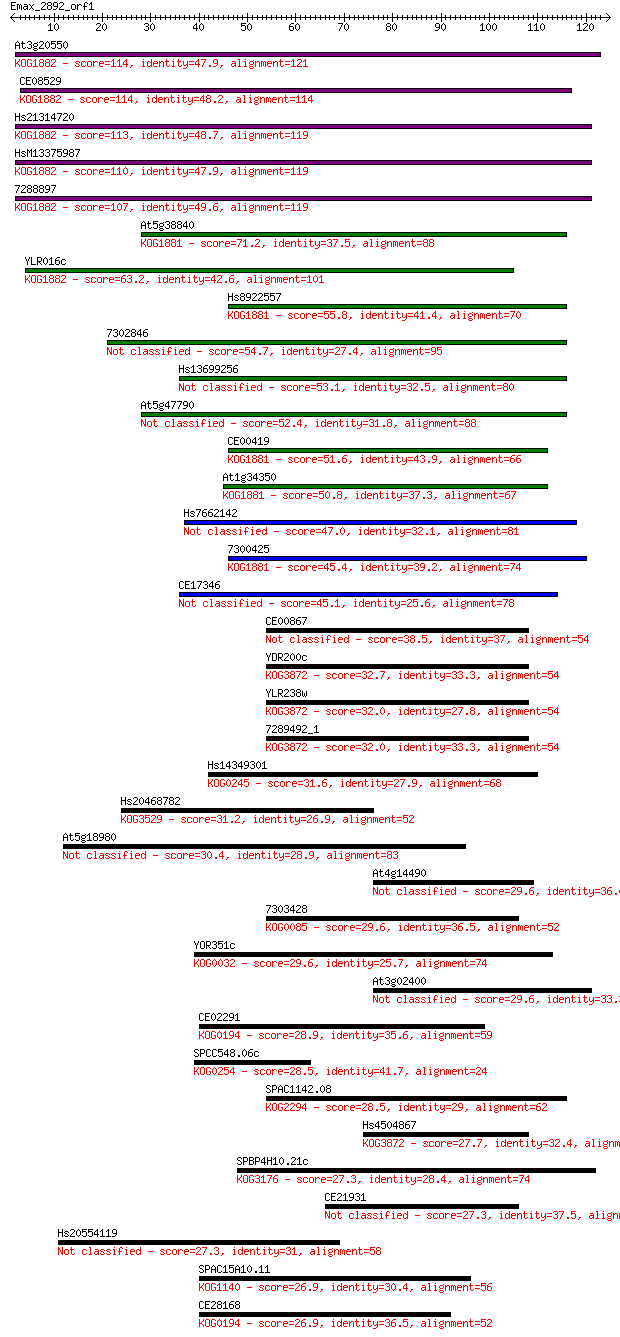
Query= Emax_2892_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
At3g20550 114 5e-26
CE08529 114 6e-26
Hs21314720 113 9e-26
HsM13375987 110 7e-25
7288897 107 8e-24
At5g38840 71.2 4e-13
YLR016c 63.2 1e-10
Hs8922557 55.8 2e-08
7302846 54.7 4e-08
Hs13699256 53.1 1e-07
At5g47790 52.4 2e-07
CE00419 51.6 4e-07
At1g34350 50.8 6e-07
Hs7662142 47.0 9e-06
7300425 45.4 3e-05
CE17346 45.1 3e-05
CE00867 38.5 0.003
YDR200c 32.7 0.16
YLR238w 32.0 0.31
7289492_1 32.0 0.32
Hs14349301 31.6 0.41
Hs20468782 31.2 0.50
At5g18980 30.4 0.86
At4g14490 29.6 1.3
7303428 29.6 1.4
YOR351c 29.6 1.5
At3g02400 29.6 1.6
CE02291 28.9 2.7
SPCC548.06c 28.5 3.0
SPAC1142.08 28.5 3.2
Hs4504867 27.7 5.5
SPBP4H10.21c 27.3 7.2
CE21931 27.3 7.3
Hs20554119 27.3 7.9
SPAC15A10.11 26.9 8.6
CE28168 26.9 10.0
> At3g20550
Length=314
Score = 114 bits (285), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 58/131 (44%), Positives = 83/131 (63%), Gaps = 20/131 (15%)
Query 2 KKWRLYLFKKENRKSAASADEEP-TKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAI 60
++WRLY+FK D EP + L +HR YLFG++ R+ DI HP+ SKQHA+
Sbjct 194 ERWRLYVFK----------DGEPLNEPLCLHRQSCYLFGRERRIADIPTDHPSCSKQHAV 243
Query 61 IQFRKTIK----GIL-----PYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSR 111
IQ+R+ K G++ PY++DL STN +Y+N+ ++ RY EL E DT++FG SSR
Sbjct 244 IQYREMEKEKPDGMMGKQVKPYIMDLGSTNKTYINESPIEPQRYYELFEKDTIKFGNSSR 303
Query 112 EFVLLHSGSVD 122
E+VLLH S +
Sbjct 304 EYVLLHENSAE 314
> CE08529
Length=299
Score = 114 bits (284), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 55/123 (44%), Positives = 80/123 (65%), Gaps = 19/123 (15%)
Query 3 KWRLYLFKKENRKSAASADEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQ 62
+WRLY FK EE +VL+IHR YL G+D ++ DI + HP+ SKQHA++Q
Sbjct 165 RWRLYPFK----------GEESLQVLYIHRQSAYLIGRDHKIADIPVDHPSCSKQHAVLQ 214
Query 63 FRKT---------IKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSREF 113
FR + I+PY+IDL S NG++LN+K+++ RYIEL+E D L+FG S+RE+
Sbjct 215 FRSMPFTRDDGTKARRIMPYIIDLGSGNGTFLNEKKIEPQRYIELQEKDMLKFGFSTREY 274
Query 114 VLL 116
V++
Sbjct 275 VVM 277
> Hs21314720
Length=396
Score = 113 bits (282), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 81/128 (63%), Gaps = 19/128 (14%)
Query 2 KKWRLYLFKKENRKSAASADEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAII 61
K+WRLY FK ++E V++IHR YL G+ R+ DI + HP+ SKQHA+
Sbjct 257 KRWRLYPFK----------NDEVLPVMYIHRQSAYLLGRHRRIADIPIDHPSCSKQHAVF 306
Query 62 QFR--------KTI-KGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSRE 112
Q+R T+ + + PY+IDL S NG++LN+KR++ RY EL+E D L+FG SSRE
Sbjct 307 QYRLVEYTRADGTVGRRVKPYIIDLGSGNGTFLNNKRIEPQRYYELKEKDVLKFGFSSRE 366
Query 113 FVLLHSGS 120
+VLLH S
Sbjct 367 YVLLHESS 374
> HsM13375987
Length=396
Score = 110 bits (275), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 57/128 (44%), Positives = 80/128 (62%), Gaps = 19/128 (14%)
Query 2 KKWRLYLFKKENRKSAASADEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAII 61
K+WRLY FK ++E V++IHR YL G+ R+ DI + HP+ SKQHA+
Sbjct 257 KRWRLYPFK----------NDEVLPVMYIHRQSAYLLGRHRRIADIPIDHPSCSKQHAVF 306
Query 62 QFR--------KTI-KGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSRE 112
Q+R T+ + + PY+IDL S NG++LN+KR++ RY EL+E D L+FG S RE
Sbjct 307 QYRLVEYTRADGTVGRRVKPYIIDLGSGNGTFLNNKRIEPQRYYELKEKDVLKFGFSIRE 366
Query 113 FVLLHSGS 120
+VLLH S
Sbjct 367 YVLLHESS 374
> 7288897
Length=421
Score = 107 bits (266), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 81/128 (63%), Gaps = 19/128 (14%)
Query 2 KKWRLYLFKKENRKSAASADEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAII 61
++WRLY FK E +A PT LHIHR +L G+D +VVD+ + HP+ SKQHA +
Sbjct 279 RRWRLYPFKGE---TAL-----PT--LHIHRQSCFLVGRDRKVVDLAVDHPSCSKQHAAL 328
Query 62 QFR---------KTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSRE 112
Q+R K + YLIDL+S NG++LN+K++D +Y EL E D ++FG SSRE
Sbjct 329 QYRLVPFEREDGSHGKRVRLYLIDLDSANGTFLNNKKIDARKYYELIEKDVIKFGFSSRE 388
Query 113 FVLLHSGS 120
+VLLH S
Sbjct 389 YVLLHENS 396
> At5g38840
Length=730
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 57/88 (64%), Gaps = 4/88 (4%)
Query 28 LHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLND 87
L +++ YLFG+D + D L HP+IS+ HA+IQ++++ Y+ DL ST+G+ +N
Sbjct 112 LDVYKKGAYLFGRD-GICDFALEHPSISRFHAVIQYKRSGAA---YIFDLGSTHGTTVNK 167
Query 88 KRVDTARYIELREGDTLRFGKSSREFVL 115
+VD +++L GD +RFG S+R ++
Sbjct 168 NKVDKKVFVDLNVGDVIRFGGSTRLYIF 195
> YLR016c
Length=204
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 43/116 (37%), Positives = 65/116 (56%), Gaps = 22/116 (18%)
Query 4 WRLYLFKKENRKSAASADEEPTKVLHIHRSMNYLFGK----------DER----VVDILL 49
+ L +++K ++ D+ P K ++ YL G+ D+R V DI +
Sbjct 78 YELVIYRKNDK------DKGPWKRYDLNGRSCYLVGRELGHSLDTDLDDRTEIVVADIGI 131
Query 50 RHPTISKQHAIIQFRKTIKGIL-PYLIDLESTNGSYLNDKRVDTARYIELREGDTL 104
T SKQH +IQFR ++GIL Y++DL+S+NG+ LN+ + ARYIELR GD L
Sbjct 132 PEETSSKQHCVIQFR-NVRGILKCYVMDLDSSNGTCLNNVVIPGARYIELRSGDVL 186
> Hs8922557
Length=796
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 44/77 (57%), Gaps = 7/77 (9%)
Query 46 DILLRHPTISKQHAIIQFRKT-------IKGILPYLIDLESTNGSYLNDKRVDTARYIEL 98
D+ L HP++S+ HA++Q R + G YL DL ST+G++LN R+ Y +
Sbjct 198 DVCLEHPSVSRYHAVLQHRASGPDGECDSNGPGFYLYDLGSTHGTFLNKTRIPPRTYCRV 257
Query 99 REGDTLRFGKSSREFVL 115
G +RFG S+R F+L
Sbjct 258 HVGHVVRFGGSTRLFIL 274
> 7302846
Length=383
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 54/95 (56%), Gaps = 2/95 (2%)
Query 21 DEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLEST 80
D++ + L + YLFG++ ++ D + H + S+ H+ + K + + YL+DL ST
Sbjct 25 DDKLVQKLMVDEKRCYLFGRNSQMNDFCIDHASCSRVHSAFVYHKHLN--IAYLVDLGST 82
Query 81 NGSYLNDKRVDTARYIELREGDTLRFGKSSREFVL 115
+G+++ R++ + +L+ T FG S+R ++L
Sbjct 83 HGTFIGTLRLEAHKPTQLQINSTFHFGASTRNYIL 117
> Hs13699256
Length=351
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 48/80 (60%), Gaps = 2/80 (2%)
Query 36 YLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARY 95
YLFG++ + D + H + S+ HA + + K +K + +LIDL ST+G++L R++ +
Sbjct 49 YLFGRNPDLCDFTIDHQSCSRVHAALVYHKHLKRV--FLIDLNSTHGTFLGHIRLEPHKP 106
Query 96 IELREGDTLRFGKSSREFVL 115
++ T+ FG S+R + L
Sbjct 107 QQIPIDSTVSFGASTRAYTL 126
> At5g47790
Length=369
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 54/88 (61%), Gaps = 4/88 (4%)
Query 28 LHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLND 87
+H+ R ++FG+ + D +L H ++S+QHA + K G + ++IDL S +G+++ +
Sbjct 80 IHLDR-RRHIFGRQHQTCDFVLDHQSVSRQHAAVVPHKN--GSI-FVIDLGSAHGTFVAN 135
Query 88 KRVDTARYIELREGDTLRFGKSSREFVL 115
+R+ +EL G +LRF S+R ++L
Sbjct 136 ERLTKDTPVELEVGQSLRFAASTRIYLL 163
> CE00419
Length=710
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 29/71 (40%), Positives = 45/71 (63%), Gaps = 7/71 (9%)
Query 46 DILLRHPTISKQHAIIQF-----RKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELRE 100
D+L+ HP+IS+ H I+Q+ KT KG ++ +L ST+GS +N KR+ +YI R
Sbjct 118 DLLMEHPSISRYHCILQYGNDKMSKTGKGW--HIFELGSTHGSRMNKKRLPPKQYIRTRV 175
Query 101 GDTLRFGKSSR 111
G +FG+S+R
Sbjct 176 GFIFQFGESTR 186
> At1g34350
Length=1587
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 43/67 (64%), Gaps = 2/67 (2%)
Query 45 VDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTL 104
DILL HP+IS+ H I+ + + + ++ DL S +G+++ D R++ +E+ EGDT+
Sbjct 182 CDILLTHPSISRFHLEIRSISSRQKL--FVTDLSSVHGTWVRDLRIEPHGCVEVEEGDTI 239
Query 105 RFGKSSR 111
R G S+R
Sbjct 240 RIGGSTR 246
> Hs7662142
Length=1584
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 46/81 (56%), Gaps = 4/81 (4%)
Query 37 LFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYI 96
G+D+ +++L+ ++ KQHA+I + + L + DL S NG+++ND R+ YI
Sbjct 24 FVGRDD--CELMLQSRSVDKQHAVINYDASTDEHL--VKDLGSLNGTFVNDVRIPEQTYI 79
Query 97 ELREGDTLRFGKSSREFVLLH 117
L+ D LRFG + F ++
Sbjct 80 TLKLEDKLRFGYDTNLFTVVQ 100
> 7300425
Length=726
Score = 45.4 bits (106), Expect = 3e-05, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 46/97 (47%), Gaps = 23/97 (23%)
Query 46 DILLRHPTISKQHAIIQFRKTI--------------------KGILP---YLIDLESTNG 82
D+ HPTIS+ H ++Q++ K P Y+ D+ ST+G
Sbjct 97 DVPAAHPTISRFHVVLQYKPKAPPKPETAKEDDEMEEEDEEPKNDQPEGWYIYDMGSTHG 156
Query 83 SYLNDKRVDTARYIELREGDTLRFGKSSREFVLLHSG 119
++LN +RV YI +R G L+ G S+R ++L G
Sbjct 157 TFLNKQRVPPKVYIRMRVGHMLKLGGSTRVYILQGPG 193
> CE17346
Length=359
Score = 45.1 bits (105), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 44/78 (56%), Gaps = 2/78 (2%)
Query 36 YLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARY 95
Y FG++ + VD + H + S+ HA++ + ++ LID++S++G++L + R+
Sbjct 45 YYFGRNNKQVDFAVEHASCSRVHALLLYHGLLQRFA--LIDMDSSHGTFLGNVRLRPLEV 102
Query 96 IELREGDTLRFGKSSREF 113
+ + G G S+R++
Sbjct 103 VFMDPGTQFHLGASTRKY 120
> CE00867
Length=952
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 33/54 (61%), Gaps = 1/54 (1%)
Query 54 ISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
IS+ HA + G+ YL++ S NG+Y+ND+R+ + L+ GDT++FG
Sbjct 65 ISRCHARVHHTNQ-DGVEEYLVEDISENGTYINDRRLSKDKREILKSGDTIKFG 117
> YDR200c
Length=604
Score = 32.7 bits (73), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 32/54 (59%), Gaps = 3/54 (5%)
Query 54 ISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
+S+ HA + T I Y+ DL+S+NG+++N ++ +EL+ GDT+ G
Sbjct 223 LSRNHACLSCDPTSGKI--YIRDLKSSNGTFVNGVKI-RQNDVELKVGDTVDLG 273
> YLR238w
Length=478
Score = 32.0 bits (71), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 33/54 (61%), Gaps = 3/54 (5%)
Query 54 ISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
+S+ HA++ + Y+ DL+S+NG+++N +R+ + +E++ GD + G
Sbjct 142 LSRNHALLSCDPLTGKV--YIRDLKSSNGTFINGQRIG-SNDVEIKVGDVIDLG 192
> 7289492_1
Length=327
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 34/54 (62%), Gaps = 4/54 (7%)
Query 54 ISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
+S+ HAI+ + T G ++ D +S+NG+++ND ++ + EL GD ++FG
Sbjct 258 LSRNHAILWY--TPDGKF-WVKDTKSSNGTFINDNKLGS-DPAELHYGDIVKFG 307
> Hs14349301
Length=1770
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query 42 ERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREG 101
ER DI+L I ++H I + ++ G + ++ + +Y+N KRV ++ ++LR G
Sbjct 518 ERRQDIVLSGAHIKEEHCIFRSERSNSGEVIVTLEPCERSETYVNGKRV--SQPVQLRSG 575
Query 102 DTLRFGKS 109
+ + GK+
Sbjct 576 NRIIMGKN 583
> Hs20468782
Length=1034
Score = 31.2 bits (69), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 30/52 (57%), Gaps = 3/52 (5%)
Query 24 PTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLI 75
PTK L H S+ Y ++RV++ L+ +++ A++Q+ K ++ + Y +
Sbjct 209 PTKTLQEHPSLAYC---EDRVIEHYLKIKGLTRGQAVVQYMKIVEALPTYGV 257
> At5g18980
Length=835
Score = 30.4 bits (67), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 5/85 (5%)
Query 12 ENRKSAASADEE--PTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIKG 69
EN +S+ A +E KV H H +++Y F + + ++L+ ++K H +G
Sbjct 436 ENTRSSGEAPDEIGEKKVFHDH-NLHYDFWRFNNLGLLILKK--LAKDHDNCGKLGNTRG 492
Query 70 ILPYLIDLESTNGSYLNDKRVDTAR 94
+LP +ID + + L D+ D AR
Sbjct 493 LLPKIIDFTHADENLLRDENADIAR 517
> At4g14490
Length=386
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 76 DLESTNGSYLNDKRVDTARYIELREGDTLRFGK 108
DL S+NG+ LN +D + L +GD ++ G+
Sbjct 64 DLGSSNGTLLNSNALDPETSVNLGDGDVIKLGE 96
> 7303428
Length=654
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 54 ISKQHAIIQFRKTIKGILPYLIDLESTNGSYLND-KRVDTARYIELREGDTLR 105
+ + I++ R GI+ Y DLE SYL+D R++ A Y+ E D LR
Sbjct 186 LPTEQDILRVRVPTTGIIEYPFDLEEIRFSYLSDLARIEQADYLP-TEQDILR 237
> YOR351c
Length=497
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query 39 GKDERVVDILLRHPTISKQHAIIQFRKTIKGILP--YLIDLESTNGSYLNDKRVDTARYI 96
G++++ ++L +P+IS H + + +P Y+ D S NG+YLN + +
Sbjct 50 GRNDKECQLVLTNPSISSVHCVFWCVFFDEDSIPMFYVKDC-SLNGTYLNGLLLKRDKTY 108
Query 97 ELREGDTLRFGKSSRE 112
L+ D + + S E
Sbjct 109 LLKHCDVIELSQGSEE 124
> At3g02400
Length=585
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 76 DLESTNGSYLNDKRVDTARYIELREGDTLRFGKSSREFVLLHSGS 120
DL S+NG+ LN +D+ + L GD ++ G+ + +L++ GS
Sbjct 64 DLGSSNGTILNSDTIDSDTPVNLSHGDEIKLGEYT--SILVNFGS 106
> CE02291
Length=414
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 28/59 (47%), Gaps = 7/59 (11%)
Query 40 KDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIEL 98
K+ RV+ L HP I K H I LPYL+ LE NG + D+ D I++
Sbjct 166 KEARVMQ-LYDHPNIVKFHGFIL------DDLPYLLVLEYCNGGAVEDRLRDKGEKIKV 217
> SPCC548.06c
Length=547
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 39 GKDERVVDILLRHPTISKQHAIIQ 62
GKDE +DI+ ++ + ++H IIQ
Sbjct 210 GKDEEALDIMCKNNVLPREHEIIQ 233
> SPAC1142.08
Length=743
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 34/65 (52%), Gaps = 9/65 (13%)
Query 54 ISKQHAIIQFRKTIKGILP---YLIDLESTNGSYLNDKRVDTARYIELREGDTLRFGKSS 110
IS+QHA I + P + I + NG++++ + V+ + + LR G ++ G+ S
Sbjct 61 ISRQHAKIFYS------FPNQRFEISVMGKNGAFVDGEFVERGKSVPLRSGTRVQIGQIS 114
Query 111 REFVL 115
F+L
Sbjct 115 FSFLL 119
> Hs4504867
Length=485
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 74 LIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
++D +S NG +LN R++ R + +GD ++ G
Sbjct 76 IMDNKSLNGVWLNRARLEPLRVYSIHQGDYIQLG 109
> SPBP4H10.21c
Length=214
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 1/74 (1%)
Query 48 LLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLRFG 107
LLRH I +H + F + + + L D + N S + +DTA + + E F
Sbjct 122 LLRHQQIVHRHYMDSFLREVPAKMNKLDD-KVGNLSMVASPDMDTAVFCVVNESVEENFR 180
Query 108 KSSREFVLLHSGSV 121
S E++ L G V
Sbjct 181 VSENEYITLDKGDV 194
> CE21931
Length=932
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 66 TIKGILPYLIDLESTNGSYLNDKRVDTARYIELREGDTLR 105
TI I L +L +++ D+ V+TA YI R G T R
Sbjct 627 TIAAIRNGLANLNGNMETFVRDQGVETAEYISQRVGSTTR 666
> Hs20554119
Length=367
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 27/58 (46%), Gaps = 0/58 (0%)
Query 11 KENRKSAASADEEPTKVLHIHRSMNYLFGKDERVVDILLRHPTISKQHAIIQFRKTIK 68
K N KS +E V ++ S Y +D+ I + P+ S HAIIQ + I+
Sbjct 27 KANGKSTVWKRQEDPPVTYVLNSPLYCVSRDQMRSKISVFLPSASSPHAIIQPKLKIR 84
> SPAC15A10.11
Length=2052
Score = 26.9 bits (58), Expect = 8.6, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Query 40 KDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDK---RVDTARY 95
K E V+ + L+H + H ++ F + P + +ST GS L D+ R+ +A+Y
Sbjct 176 KPEEVIPLELQHSIRTTIHILLDFILDVFSCSPVNLKAQSTVGSILADEEASRLSSAKY 234
> CE28168
Length=395
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 7/52 (13%)
Query 40 KDERVVDILLRHPTISKQHAIIQFRKTIKGILPYLIDLESTNGSYLNDKRVD 91
K+ RV+ L HP I ++F I +PYL+ LE NG + DK V+
Sbjct 166 KEARVMQ-LYDHPNI------VKFYGYIVDDIPYLLVLELCNGGSVEDKLVE 210
Lambda K H
0.320 0.137 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40