bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2893_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
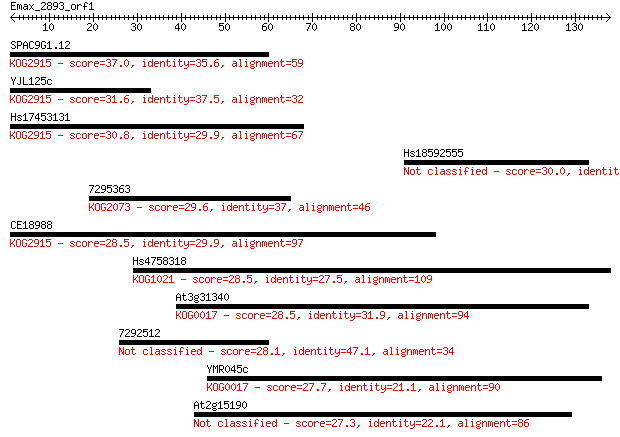
SPAC9G1.12 37.0 0.011
YJL125c 31.6 0.41
Hs17453131 30.8 0.72
Hs18592555 30.0 1.3
7295363 29.6 1.9
CE18988 28.5 3.8
Hs4758318 28.5 4.0
At3g31340 28.5 4.3
7292512 28.1 4.9
YMR045c 27.7 6.2
At2g15190 27.3 9.3
> SPAC9G1.12
Length=364
Score = 37.0 bits (84), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 3/59 (5%)
Query 1 SPCVEQLHKTFAAAQELGFQDFEAFEILAKPWGACISRPAPGQKKDENPKRKRDASSQR 59
SPC+EQ+ + A +ELG+ D E E+ K W A SR DE R ++ +R
Sbjct 213 SPCIEQIQHSAEALRELGWCDIEMIEVDYKQWAARKSRIV---HIDEAIDRLKEVKRRR 268
> YJL125c
Length=383
Score = 31.6 bits (70), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 1 SPCVEQLHKTFAAAQELGFQDFEAFEILAKPW 32
SPC+EQ+ KT ++ G+ D E EI + +
Sbjct 230 SPCIEQVDKTLDVLEKYGWTDVEMVEIQGRQY 261
> Hs17453131
Length=289
Score = 30.8 bits (68), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 3/70 (4%)
Query 1 SPCVEQLHKTFAAAQELGFQDFEAFEILAKPWGA-CISRPAPGQKKDENPKRKRDASSQR 59
SPC+EQ+ +T A GF + E+L + + +S P P + D S R
Sbjct 207 SPCIEQVQRTCQALAARGFSELSTLEVLPQVYNVRTVSLPPPDLGTGTDGPAGSDTSPFR 266
Query 60 --QPEEEVYG 67
P +E G
Sbjct 267 SGTPMKEAVG 276
> Hs18592555
Length=185
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 28/47 (59%), Gaps = 7/47 (14%)
Query 91 LQQPFSQSARWCSADLSARGDVAPSP----LKKASEIH-SAEPSRIS 132
L+QPFS S WCS D R ++PSP ++A E+ +PSRI+
Sbjct 105 LEQPFSASQNWCSRD--GRRAISPSPPAFSTREACEMTLPPKPSRIT 149
> 7295363
Length=962
Score = 29.6 bits (65), Expect = 1.9, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 19 FQDFEA-FEILAKPWGACISRPAPGQKKDENPKRKRDASSQRQPEEE 64
F DF+A F G IS P PG++++E+ ++++Q PEEE
Sbjct 771 FADFDAHFSTFGSDLGMPISAP-PGEEEEEDSSPAPESATQVNPEEE 816
> CE18988
Length=312
Score = 28.5 bits (62), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 47/106 (44%), Gaps = 30/106 (28%)
Query 1 SPCVEQLHKTFAAAQELGFQDFEAFEILAKPWGACISRPAPGQKKDENPKRKRDASSQRQ 60
SPC+EQ+ + A GF E E++ P++K+ S RQ
Sbjct 211 SPCLEQVQRACLAMAAAGFVSIETIELV--------------------PEQKKIVSQHRQ 250
Query 61 PEEEV--YGDA--QERQ---DGQ--RQTATSFPALLSHFLQQPFSQ 97
EE GDA +ER+ DG Q++++ P ++ + P+SQ
Sbjct 251 TLEEFEELGDAYPKERKRNVDGTIVEQSSSAIPR-ITIAIVYPYSQ 295
> Hs4758318
Length=676
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 45/112 (40%), Gaps = 13/112 (11%)
Query 29 AKPW-GACISRPAPGQ--KKDENPKRKRDASSQRQPEEEVYGDAQERQDGQRQTATSFPA 85
A PW G C P PGQ +++ P S +PE + Q A P
Sbjct 245 ACPWDGRCEQDPGPGQTQRQETLPNATFCLISGHRPEAA-------SRFLQALQAGCIPV 297
Query 86 LLSHFLQQPFSQSARWCSADLSARGDVAPSPLKKASEIHSAEPSRISSYHYQ 137
LLS + PFS+ W A + A + PL+ + + P+R+ + Q
Sbjct 298 LLSPRWELPFSEVIDWTKAAIVADERL---PLQVLAALQEMSPARVLALRQQ 346
> At3g31340
Length=781
Score = 28.5 bits (62), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 46/110 (41%), Gaps = 16/110 (14%)
Query 39 PAPGQKKDENPKRKRD----ASSQRQPEEEVYGDAQERQDGQRQTATSF---PALLSHFL 91
P PG+++ +++ + SQRQ +E D E G+ +T PA + L
Sbjct 473 PFPGRERQIQSRKREEYALLVESQRQQKEAQLTDVVEVLAGKEMVSTCSAIPPATIPEKL 532
Query 92 QQP--FSQSARW-------CSADLSARGDVAPSPLKKASEIHSAEPSRIS 132
P F R C DL A ++ P + K I + +PSRIS
Sbjct 533 GDPGSFVLPCRIGKSAFERCLCDLGAGVNLMPLSMSKRLGITNFKPSRIS 582
> 7292512
Length=1042
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 26 EILAKPWGACISRPAPGQKKDENPKRKRDASSQR 59
E++A P RP PGQK KRKR SS R
Sbjct 384 ELIAGPRYRLGKRPLPGQKSGSPIKRKRVTSSLR 417
> YMR045c
Length=1755
Score = 27.7 bits (60), Expect = 6.2, Method: Composition-based stats.
Identities = 19/90 (21%), Positives = 41/90 (45%), Gaps = 6/90 (6%)
Query 46 DENPKRKRDASSQRQPEEEVYGDAQERQDGQRQTATSFPALLSHFLQQPFSQSARWCSAD 105
+E + KR+ S+ R+ + E++D + T T+ P ++ Q+P + +R A
Sbjct 348 EEQQESKRNKSTHRRSPSD------EKKDSRTYTNTTKPKSITRNSQKPNNSQSRTARAH 401
Query 106 LSARGDVAPSPLKKASEIHSAEPSRISSYH 135
+ + +P P + EP ++ + H
Sbjct 402 NVSTFNNSPGPDNDLIRGSTTEPIQLKNTH 431
> At2g15190
Length=775
Score = 27.3 bits (59), Expect = 9.3, Method: Composition-based stats.
Identities = 19/87 (21%), Positives = 41/87 (47%), Gaps = 8/87 (9%)
Query 43 QKKDENPKRKR-DASSQRQPEEEVYGDAQERQDGQRQTATSFPALLSHFLQQPFSQSARW 101
Q+ +E+ +RK+ D +RQPE E G + + + ++ +++P +Q +
Sbjct 372 QRSEEDEERKQEDEGVERQPEAEEEGGLERKAENDNES-------FEDSIREPNTQYGSY 424
Query 102 CSADLSARGDVAPSPLKKASEIHSAEP 128
D + + DV +++S+ S P
Sbjct 425 PGDDENTQRDVGDEVAEESSKDKSPTP 451
Lambda K H
0.313 0.126 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40