bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2911_orf1
Length=107
Score E
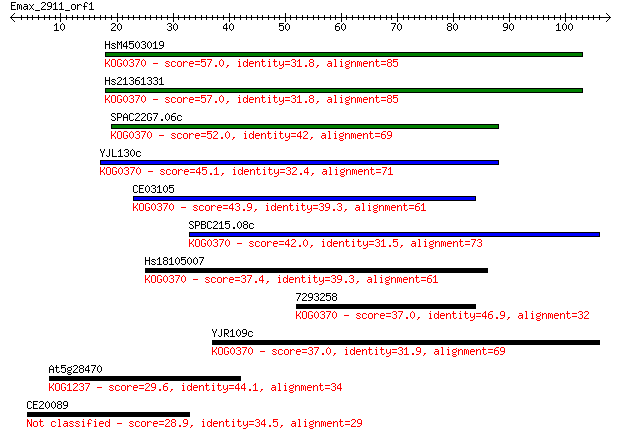
Sequences producing significant alignments: (Bits) Value
HsM4503019 57.0 7e-09
Hs21361331 57.0 7e-09
SPAC22G7.06c 52.0 3e-07
YJL130c 45.1 4e-05
CE03105 43.9 8e-05
SPBC215.08c 42.0 2e-04
Hs18105007 37.4 0.007
7293258 37.0 0.010
YJR109c 37.0 0.010
At5g28470 29.6 1.4
CE20089 28.9 2.4
> HsM4503019
Length=1500
Score = 57.0 bits (136), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query 18 RETHNSGRLTAKQAIEKGLVEMVINVPGQTNHRAISSGYLIRRAAIDAGVPLLTDLKVAA 77
+E N + ++ I G +++VIN+P N + + Y+IRR A+D+G+PLLT+ +V
Sbjct 1413 QEGQNPSLSSIRKLIRDGSIDLVINLP-NNNTKFVHDNYVIRRTAVDSGIPLLTNFQVTK 1471
Query 78 LFVESIAKKIYREQERQRFWEVRAW 102
LF E++ K R+ + + + R +
Sbjct 1472 LFAEAVQKS--RKVDSKSLFHYRQY 1494
> Hs21361331
Length=1500
Score = 57.0 bits (136), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query 18 RETHNSGRLTAKQAIEKGLVEMVINVPGQTNHRAISSGYLIRRAAIDAGVPLLTDLKVAA 77
+E N + ++ I G +++VIN+P N + + Y+IRR A+D+G+PLLT+ +V
Sbjct 1413 QEGQNPSLSSIRKLIRDGSIDLVINLP-NNNTKFVHDNYVIRRTAVDSGIPLLTNFQVTK 1471
Query 78 LFVESIAKKIYREQERQRFWEVRAW 102
LF E++ K R+ + + + R +
Sbjct 1472 LFAEAVQKS--RKVDSKSLFHYRQY 1494
> SPAC22G7.06c
Length=2244
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 43/73 (58%), Gaps = 6/73 (8%)
Query 19 ETHNSGRLTAKQAIEKGLVEMVINVPGQTNHR----AISSGYLIRRAAIDAGVPLLTDLK 74
E +N L+A A +++M IN+P +R ISSGY RR AID VPL+T++K
Sbjct 1451 EANNEYSLSAHLA--NNMIDMYINLPSNNRYRRPANYISSGYKSRRLAIDYSVPLVTNVK 1508
Query 75 VAALFVESIAKKI 87
A L +E+I + +
Sbjct 1509 CAKLMIEAICRNL 1521
> YJL130c
Length=2214
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 39/75 (52%), Gaps = 4/75 (5%)
Query 17 KRETHNSGRLTAKQAIEKGLVEMVINVPGQTNHR----AISSGYLIRRAAIDAGVPLLTD 72
K + + Q + +++ IN+P R +S GY RR A+D VPL+T+
Sbjct 1415 KDDDDQKSEYSLTQHLANNEIDLYINLPSANRFRRPASYVSKGYKTRRLAVDYSVPLVTN 1474
Query 73 LKVAALFVESIAKKI 87
+K A L +E+I++ I
Sbjct 1475 VKCAKLLIEAISRNI 1489
> CE03105
Length=2198
Score = 43.9 bits (102), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 38/66 (57%), Gaps = 5/66 (7%)
Query 23 SGRLTAKQAIEKGLVEMVINVP--GQTNHRAI---SSGYLIRRAAIDAGVPLLTDLKVAA 77
SG + + +E +VIN+P G +R + GY RR AID G+PL+TD+K A
Sbjct 1387 SGTRSVVEFLENKEFHLVINLPIRGSGAYRVSAFRTHGYKTRRMAIDNGIPLITDIKCAK 1446
Query 78 LFVESI 83
F++++
Sbjct 1447 TFIQAL 1452
> SPBC215.08c
Length=1160
Score = 42.0 bits (97), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 40/75 (53%), Gaps = 2/75 (2%)
Query 33 EKGLVEMVINVPGQTNHRAISSGYLIRRAAIDAGVPLLTDLKVAALFVESIAKKI--YRE 90
EK + V N+ + Y++RR A+D V L+ D+ A LFVES+ +K+
Sbjct 1080 EKYNIRSVFNLASARGKNVLDQDYVMRRNAVDFNVTLINDVNCAKLFVESLKEKLPSVLS 1139
Query 91 QERQRFWEVRAWDEY 105
++++ EV+ W E+
Sbjct 1140 EKKEMPSEVKRWSEW 1154
> Hs18105007
Length=2225
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 35/66 (53%), Gaps = 6/66 (9%)
Query 25 RLTAKQAIEKGLVEMVINVP-----GQTNHRAISSGYLIRRAAIDAGVPLLTDLKVAALF 79
R +Q EK E+VIN+ G+ ++ GY RR A D VPL+ D+K LF
Sbjct 1378 RSILEQLAEKNF-ELVINLSMRGAGGRRLSSFVTKGYRTRRLAADFSVPLIIDIKCTKLF 1436
Query 80 VESIAK 85
VE++ +
Sbjct 1437 VEALGQ 1442
> 7293258
Length=2189
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 52 ISSGYLIRRAAIDAGVPLLTDLKVAALFVESI 83
++ GY RR A+D +PL+TD+K L VES+
Sbjct 1389 MTHGYRTRRLAVDYSIPLVTDVKCTKLLVESM 1420
> YJR109c
Length=1118
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 12/78 (15%)
Query 37 VEMVINVPGQTNHRAISSGYLIRRAAIDAGVPLLTDLKVAALFVESIAKKIYREQERQRF 96
++ V N+ + Y++RR AID +PL + + A LF + + KI E+ +
Sbjct 1038 IKAVFNLASKRAESTDDVDYIMRRNAIDFAIPLFNEPQTALLFAKCLKAKI---AEKIKI 1094
Query 97 W---------EVRAWDEY 105
EVR+WDE+
Sbjct 1095 LESHDVIVPPEVRSWDEF 1112
> At5g28470
Length=561
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 8 FETQIIPVCKRETHNSGRLTAKQAIEKGLVEMVI 41
+E IIP+ K+ T RLT K IE G+V +I
Sbjct 386 YECVIIPIVKQITGRKKRLTLKHRIEIGIVMGII 419
> CE20089
Length=334
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 10/29 (34%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 4 PFKAFETQIIPVCKRETHNSGRLTAKQAI 32
P++ + Q+ P CKR +S TA Q++
Sbjct 301 PYREYTQQLFPFCKRLDFSSSNQTAPQSV 329
Lambda K H
0.321 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40