bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2913_orf2
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
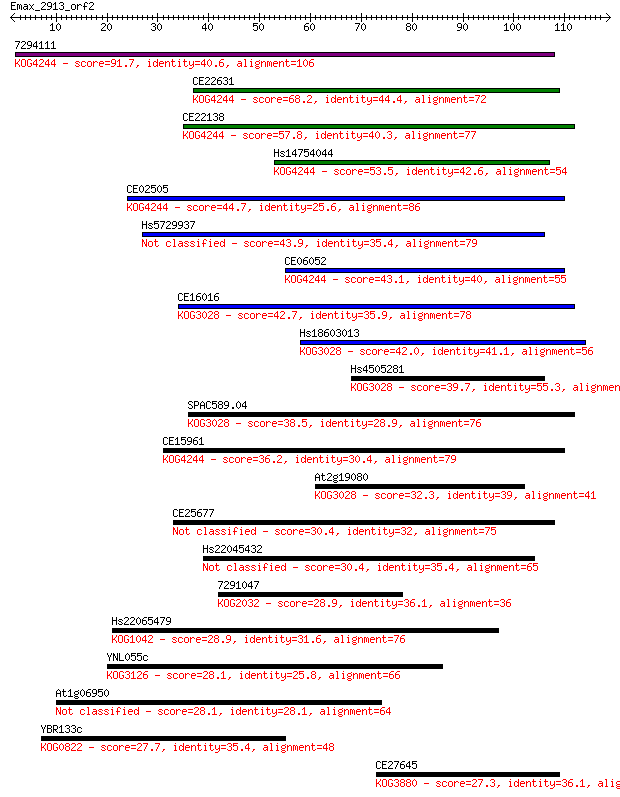
7294111 91.7 3e-19
CE22631 68.2 4e-12
CE22138 57.8 5e-09
Hs14754044 53.5 9e-08
CE02505 44.7 4e-05
Hs5729937 43.9 7e-05
CE06052 43.1 1e-04
CE16016 42.7 2e-04
Hs18603013 42.0 2e-04
Hs4505281 39.7 0.001
SPAC589.04 38.5 0.003
CE15961 36.2 0.014
At2g19080 32.3 0.24
CE25677 30.4 0.82
Hs22045432 30.4 0.86
7291047 28.9 2.3
Hs22065479 28.9 2.4
YNL055c 28.1 3.9
At1g06950 28.1 4.3
YBR133c 27.7 4.8
CE27645 27.3 6.2
> 7294111
Length=415
Score = 91.7 bits (226), Expect = 3e-19, Method: Composition-based stats.
Identities = 43/107 (40%), Positives = 65/107 (60%), Gaps = 1/107 (0%)
Query 2 WIVRWWRYNHPQEFLDTAQLDIKRTFNSRFPNSILNFVFKIGF-RKHLKDAVGYGVGRFT 60
WI+ +WR +P L +++++ R PNSILNF FKI F RK K +G+G +
Sbjct 207 WIIFYWRAKYPDNVLKGYKVNLQHALGLRLPNSILNFFFKITFGRKGTKKLKAHGIGVHS 266
Query 61 EAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHLAQLLYV 107
E+ + ++DL LS +L K +FFG EP +LD+ AF+ L+QL Y+
Sbjct 267 AEEIEEFGKDDLKVLSEMLDCKPFFFGDEPTTLDVVAFAVLSQLHYL 313
> CE22631
Length=269
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 37 NFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDIT 96
NF+ K +K A G+G F+ EV+ A++DL A+ST LG K Y FG ++D+T
Sbjct 150 NFIVKSFTKKVRGRAAAQGMGTFSREEVLDQAKKDLDAISTQLGDKPYLFGSSIKTIDVT 209
Query 97 AFSHLAQLLYVP 108
AF+HLA+L+Y P
Sbjct 210 AFAHLAELIYTP 221
> CE22138
Length=321
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 43/82 (52%), Gaps = 5/82 (6%)
Query 35 ILNFVFK-IGFRKH----LKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKE 89
I F+FK IG R + L+ G +G + ++I EDL A+S LG K YF G +
Sbjct 185 IFGFLFKLIGSRTYVSALLRRITGSDIGFHSREDIINIGSEDLKAISKYLGNKHYFHGFK 244
Query 90 PHSLDITAFSHLAQLLYVPFAG 111
P +D FSHL Q+ Y P+
Sbjct 245 PTKVDACIFSHLCQIYYAPYTS 266
> Hs14754044
Length=409
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 36/54 (66%), Gaps = 0/54 (0%)
Query 53 GYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHLAQLLY 106
G+G+GRF+E E+ ++D+ +L+ LLG K Y G + +LD T F HLAQ ++
Sbjct 248 GHGIGRFSEEEIYMLMEKDMRSLAGLLGDKKYIMGPKLSTLDATVFGHLAQAMW 301
> CE02505
Length=275
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 44/86 (51%), Gaps = 1/86 (1%)
Query 24 KRTFNSRFPNSILNFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKA 83
++ N P+ + N++ K GF + + V +G+ AE + + D+ A+ +LG K
Sbjct 158 RQVSNLPLPSFLTNYLAK-GFSQTARKRVNGVLGKLDVAEQKELLRRDIRAIDDILGDKK 216
Query 84 YFFGKEPHSLDITAFSHLAQLLYVPF 109
+ FG S+D + F + + Y+P+
Sbjct 217 FLFGDRITSVDCSVFGQIGAVFYLPY 242
> Hs5729937
Length=263
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 6/82 (7%)
Query 27 FNSRFP---NSILNFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKA 83
+ S +P N IL + + ++ +K A+G+G + T +V++ + ALS LGT+
Sbjct 145 YGSPYPWPLNHILAYQKQWEVKRKMK-AIGWG--KKTLDQVLEDVDQCCQALSQRLGTQP 201
Query 84 YFFGKEPHSLDITAFSHLAQLL 105
YFF K+P LD F HL +L
Sbjct 202 YFFNKQPTELDALVFGHLYTIL 223
> CE06052
Length=287
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 55 GVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHLAQLLYVPF 109
+G F AE+ + DL + T+LG K Y FG +D T FS LA ++Y PF
Sbjct 201 AIGDFEPAELDELLHRDLRVVETILGNKKYLFGDHIAPVDATVFSQLA-VVYYPF 254
> CE16016
Length=312
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 34 SILNFVFKIGF-RKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHS 92
S L+F + + + K K A+ G+ + E+++ A L LST LG +F G +P S
Sbjct 129 SHLHFPYNLYYLEKRRKKALRLLAGK-NDTEILKEAFMALNTLSTKLGDNKFFCGNKPTS 187
Query 93 LDITAFSHLAQLLYVPFAG 111
LD F +LA LL VP
Sbjct 188 LDALVFGYLAPLLRVPLPN 206
> Hs18603013
Length=326
Score = 42.0 bits (97), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 58 RFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHLAQLLYVPFAGLK 113
R EA++ + A+E L LS LGT +FFG P +LD F LA L V F ++
Sbjct 163 REVEAQIYRDAKECLNLLSNRLGTSQFFFGDTPSTLDAYVFGFLAPLYKVRFPKVQ 218
> Hs4505281
Length=317
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/38 (55%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 68 AQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHLAQLL 105
A+E LT LS LG++ +FFG P SLD FS+LA LL
Sbjct 174 ARECLTLLSQRLGSQKFFFGDAPASLDAFVFSYLALLL 211
> SPAC589.04
Length=271
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 39/79 (49%), Gaps = 3/79 (3%)
Query 36 LNFVFKIGFRKHLKDAVGYGVGRFTEA--EVIQAAQEDLTALSTLLGTKAYFFGKE-PHS 92
LN + IG +K + + T +++ A + +ALS LLG+ +FF E P
Sbjct 158 LNIIKSIGLPSQIKRKICLQLNESTLDFDAILEDASKAFSALSELLGSDKWFFNNESPSF 217
Query 93 LDITAFSHLAQLLYVPFAG 111
LD++ F+H + ++P
Sbjct 218 LDVSLFAHAEIINHLPLKN 236
> CE15961
Length=277
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 1/79 (1%)
Query 31 FPNSILNFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEP 90
F N +L + K K++ + +G F +E+ + DL + L K + FG+E
Sbjct 167 FLNFLLMPLLKAIIGKNVYNKCQGAIGDFELSELDEILHRDLRIVENTLAKKKFLFGEEI 226
Query 91 HSLDITAFSHLAQLLYVPF 109
+ D T FS LA +Y PF
Sbjct 227 TAADATVFSQLAT-VYYPF 244
> At2g19080
Length=315
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 61 EAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSLDITAFSHL 101
E ++ + A E ALST LG + + F P SLD SH+
Sbjct 163 EKQIYKRASEAYEALSTRLGEQKFLFEDRPSSLDAFLLSHI 203
> CE25677
Length=244
Score = 30.4 bits (67), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 3/75 (4%)
Query 33 NSILNFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHS 92
+S+L FV + RK L++ T EV + A + ALS LG++ Y G P
Sbjct 142 SSVLPFVKR---RKILEELSDKDWDTKTMDEVGEQADKVFRALSAQLGSQKYLTGDLPTE 198
Query 93 LDITAFSHLAQLLYV 107
D F H+ L+ V
Sbjct 199 ADALLFGHMYTLITV 213
> Hs22045432
Length=328
Score = 30.4 bits (67), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 5/70 (7%)
Query 39 VFKIGFRKHL--KDAVGYGVGRFTEAEVIQAAQEDLTALSTLLGTKAYFFGKEPHSL--- 93
V G H+ +A Y + E V+Q QE+ T +LLG KA G E L
Sbjct 92 VKDCGISSHILQNEADSYTSNQHLECHVLQKQQEEGTQSKSLLGPKARECGSEDLGLGTS 151
Query 94 DITAFSHLAQ 103
T SHL +
Sbjct 152 KATRKSHLGK 161
> 7291047
Length=822
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Query 42 IGFRK-HLKDAVGYGVGRFTEAEVIQAAQEDLTALST 77
+G R+ H+K A Y +GRF+EA + ++ +++ ST
Sbjct 222 VGIRQDHIKQAHAYAIGRFSEALLEESGKKEEENCST 258
> Hs22065479
Length=852
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 36/78 (46%), Gaps = 4/78 (5%)
Query 21 LDIKRTFNSRFPNSILNFVFKIGFRKHLKDAVGYGVGR--FTEAEVIQAAQEDLTALSTL 78
+ +KR S P I VF I FRK LK Y +GR + +E ++ Q L+
Sbjct 189 ITLKRELPSSSPVCIQ--VFNIIFRKILKKLSMYQIGRNFYNPSEPMEIPQHKLSLWPGF 246
Query 79 LGTKAYFFGKEPHSLDIT 96
+ +YF K S D++
Sbjct 247 AISVSYFERKLLFSADVS 264
> YNL055c
Length=283
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 30/66 (45%), Gaps = 3/66 (4%)
Query 20 QLDIKRTFNSRFPNSILNFVFKIGFRKHLKDAVGYGVGRFTEAEVIQAAQEDLTALSTLL 79
Q+ K T N + PNS +N F ++L DA + +++ ++ A + L L
Sbjct 201 QVGAKATMNCKLPNSNVNIEFAT---RYLPDASSQVKAKVSDSGIVTLAYKQLLRPGVTL 257
Query 80 GTKAYF 85
G + F
Sbjct 258 GVGSSF 263
> At1g06950
Length=1016
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query 10 NHPQEFLDTAQLDIKRTFNSRFPNSILNFVFKIG-----FRKHLKDAVGYGVGRFTEAEV 64
N + ++ +L+IK+ + N L+ + + F+K + D G G F E EV
Sbjct 829 NAIETAVNQGRLNIKQIRELKEANVSLDSMIAVSLREKLFKKTVSDIFSSGTGEFDETEV 888
Query 65 IQAAQEDLT 73
Q DL+
Sbjct 889 YQTIPSDLS 897
> YBR133c
Length=827
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 22/48 (45%), Gaps = 3/48 (6%)
Query 7 WRYNHPQEFLDTAQLDIKRTFNSRFPNSILNFVFKIGFRKHLKDAVGY 54
WR+ HP DT Q + F F S LN FKI R + +G+
Sbjct 542 WRFEHPMAQKDTVQDE--DDFTVEFSQSSLN-EFKIKHRGEIHGFIGF 586
> CE27645
Length=417
Score = 27.3 bits (59), Expect = 6.2, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 73 TALSTLLGTKAYFFGKEPHSLDITAFSHLAQLLYVP 108
T ++ LLG+ +Y F EPH ++T L L++P
Sbjct 157 TGMAGLLGSFSYAFLTEPHMANLTPKVALLIQLFIP 192
Lambda K H
0.325 0.139 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40