bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2927_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
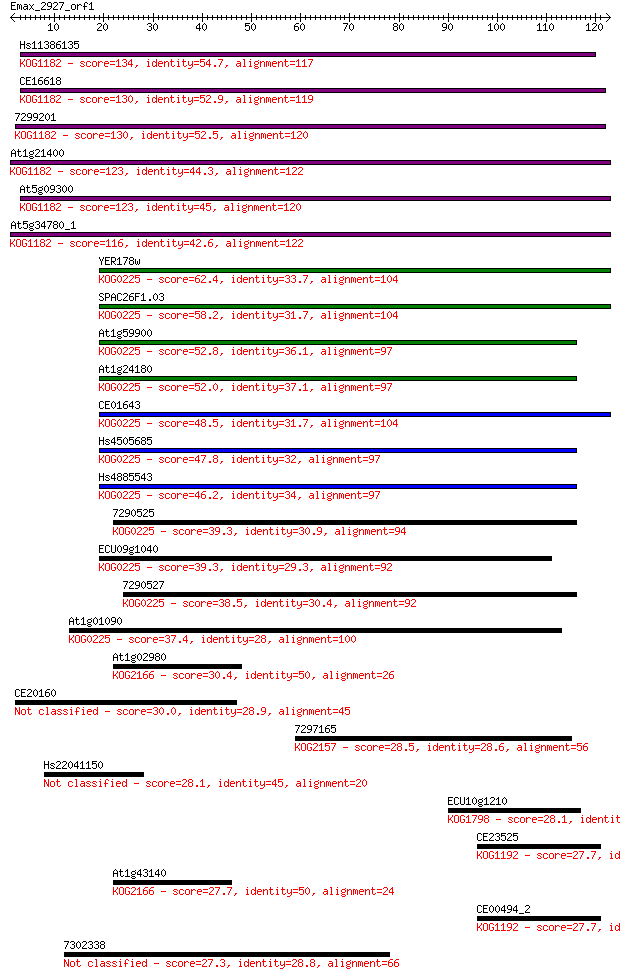
Hs11386135 134 3e-32
CE16618 130 4e-31
7299201 130 7e-31
At1g21400 123 7e-29
At5g09300 123 1e-28
At5g34780_1 116 1e-26
YER178w 62.4 2e-10
SPAC26F1.03 58.2 4e-09
At1g59900 52.8 2e-07
At1g24180 52.0 3e-07
CE01643 48.5 3e-06
Hs4505685 47.8 6e-06
Hs4885543 46.2 2e-05
7290525 39.3 0.002
ECU09g1040 39.3 0.002
7290527 38.5 0.003
At1g01090 37.4 0.006
At1g02980 30.4 0.86
CE20160 30.0 1.0
7297165 28.5 3.5
Hs22041150 28.1 4.7
ECU10g1210 28.1 4.7
CE23525 27.7 4.8
At1g43140 27.7 5.2
CE00494_2 27.7 5.8
7302338 27.3 7.9
> Hs11386135
Length=445
Score = 134 bits (338), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 64/117 (54%), Positives = 86/117 (73%), Gaps = 1/117 (0%)
Query 3 DQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQH 62
+QY GDGIA RG YGI +IR+DGND+FA + ATK+AR+ + N+P L+E MTYR+G H
Sbjct 277 EQYRGDGIAARGPGYGIMSIRVDGNDVFAVYNATKEARRRAVAENQPFLIEAMTYRIGHH 336
Query 63 STSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLE 119
STSDDSSAYR E+ W+ PI+RLR YL +GWW +E+E+ RK+SR+ ++E
Sbjct 337 STSDDSSAYRSVDEVNYWDKQD-HPISRLRHYLLSQGWWDEEQEKAWRKQSRRKVME 392
> CE16618
Length=478
Score = 130 bits (328), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 63/119 (52%), Positives = 85/119 (71%), Gaps = 2/119 (1%)
Query 3 DQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQH 62
+QY GDGIA +G +YG+ TIR+DGNDL A + ATK+AR++ L TN PVL+E MTYR+G H
Sbjct 308 EQYGGDGIAGKGPAYGLHTIRVDGNDLLAVYNATKEARRVAL-TNRPVLIEAMTYRLGHH 366
Query 63 STSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQM 121
STSDDS+AYR E++ W PI R +KY+ GWW +E+E E +KE +K +L +
Sbjct 367 STSDDSTAYRSSDEVQTWGDKD-HPITRFKKYITERGWWNEEKEMEWQKEVKKRVLTEF 424
> 7299201
Length=439
Score = 130 bits (327), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 63/121 (52%), Positives = 87/121 (71%), Gaps = 2/121 (1%)
Query 2 RDQYAGDGIAIRG-ISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVG 60
+QY GDGIA RG + YGI TIR+DG D+FA + A K AR+ +L N+PV+ E + YRVG
Sbjct 269 HEQYKGDGIAGRGPMGYGITTIRVDGTDVFAVYNAMKAAREYVLKENKPVVFEALAYRVG 328
Query 61 QHSTSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQ 120
HSTSDDS+AYRP E+E WN S PI++L++Y+ H+GW+ + E E K+ RK +L+Q
Sbjct 329 HHSTSDDSTAYRPAEEIEIWN-SVEHPISKLKRYMVHKGWFDETVENEYVKDIRKKVLKQ 387
Query 121 M 121
+
Sbjct 388 I 388
> At1g21400
Length=472
Score = 123 bits (309), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 54/122 (44%), Positives = 84/122 (68%), Gaps = 1/122 (0%)
Query 1 VRDQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVG 60
+ +Q+ DGI ++G +YGI++IR+DGND A + A + AR++ + PVL+E MTYRVG
Sbjct 303 ISEQFRSDGIVVKGQAYGIRSIRVDGNDALAVYSAVRSAREMAVTEQRPVLIEMMTYRVG 362
Query 61 QHSTSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQ 120
HSTSDDS+ YR E++ W S P+ R RK++ GWW++E+E +LR +RK +L+
Sbjct 363 HHSTSDDSTKYRAADEIQYWKMSRN-PVNRFRKWVEDNGWWSEEDESKLRSNARKQLLQA 421
Query 121 MK 122
++
Sbjct 422 IQ 423
> At5g09300
Length=414
Score = 123 bits (308), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 54/120 (45%), Positives = 82/120 (68%), Gaps = 1/120 (0%)
Query 3 DQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQH 62
DQ+ DG+ ++G +YGI++IR+DGND A + A AR++ + P+L+E +TYRVG H
Sbjct 247 DQFRSDGVVVKGRAYGIRSIRVDGNDALAMYSAVHTAREMAIREQRPILIEALTYRVGHH 306
Query 63 STSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQMK 122
STSDDS+ YR GE+E WN + P++R R ++ GWW+D+ E +LR +K MLE ++
Sbjct 307 STSDDSTRYRSAGEIEWWNKAR-NPLSRFRTWIESNGWWSDKTESDLRSRIKKEMLEALR 365
> At5g34780_1
Length=288
Score = 116 bits (290), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 52/122 (42%), Positives = 80/122 (65%), Gaps = 1/122 (0%)
Query 1 VRDQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVG 60
+ +Q+ DGI ++G +YGI++IR+DGND A + A AR++ + PVL+E M YRVG
Sbjct 73 ISEQFRSDGIVVKGQAYGIRSIRVDGNDALAVYSAVCSAREMAVTEQRPVLIEMMIYRVG 132
Query 61 QHSTSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQ 120
HSTSDDS+ YR E++ W S + R RK + GWW++E+E +LR +RK +L+
Sbjct 133 HHSTSDDSTKYRAADEIQYWKMSRN-SVNRFRKSVEDNGWWSEEDESKLRSNARKQLLQA 191
Query 121 MK 122
++
Sbjct 192 IQ 193
> YER178w
Length=443
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 59/104 (56%), Gaps = 1/104 (0%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
I ++++G D+ A + A+K A+ L P+++E+ TYR G HS SD + YR E++
Sbjct 292 IPGLKVNGMDILAVYQASKFAKDWCLSGKGPLVLEYETYRYGGHSMSDPGTTYRTRDEIQ 351
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQMK 122
+ PIA L+ +L G T+ E + K +RKY+ EQ++
Sbjct 352 HMRSKND-PIAGLKMHLIDLGIATEAEVKAYDKSARKYVDEQVE 394
> SPAC26F1.03
Length=409
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 59/104 (56%), Gaps = 1/104 (0%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
I + ++G D+ A A+K A++ ++ ++P+LMEF+TYR G HS SD + YR E++
Sbjct 266 IPGLLVNGMDVLAVLQASKFAKKYTVENSQPLLMEFVTYRYGGHSMSDPGTTYRSREEVQ 325
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQMK 122
A+ PI L+K++ G E + + K R + E+++
Sbjct 326 KVRAARD-PIEGLKKHIMEWGVANANELKNIEKRIRGMVDEEVR 368
> At1g59900
Length=389
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 54/101 (53%), Gaps = 10/101 (9%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
+ +++DG D FA A K A+Q L+ P+++E TYR HS SD S YR E+
Sbjct 249 VPGLKVDGMDAFAVKQACKFAKQHALEKG-PIILEMDTYRYHGHSMSDPGSTYRTRDEI- 306
Query 79 AWNASGIL----PIARLRKYLAHEGWWTDEEEEELRKESRK 115
SG+ PI R++K + T++E +++ KE RK
Sbjct 307 ----SGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRK 343
> At1g24180
Length=393
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 53/101 (52%), Gaps = 10/101 (9%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
+ +++DG D A A K A++ L N P+++E TYR HS SD S YR E+
Sbjct 253 VPGLKVDGMDALAVKQACKFAKEHALK-NGPIILEMDTYRYHGHSMSDPGSTYRTRDEI- 310
Query 79 AWNASGIL----PIARLRKYLAHEGWWTDEEEEELRKESRK 115
SG+ PI R+RK L T++E +++ KE RK
Sbjct 311 ----SGVRQVRDPIERVRKLLLTHDIATEKELKDMEKEIRK 347
> CE01643
Length=397
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 52/104 (50%), Gaps = 1/104 (0%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
+ I +DG D+ A ATK A++ P++ME TYR HS SD ++YR E++
Sbjct 243 VPGIWVDGMDILAVREATKWAKEYCDSGKGPLMMEMATYRYHGHSMSDPGTSYRTREEIQ 302
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRKYMLEQMK 122
+ PI + + T+EE + + KE RK + E +K
Sbjct 303 EVRKTRD-PITGFKDRIITSSLATEEELKAIDKEVRKEVDEALK 345
> Hs4505685
Length=390
Score = 47.8 bits (112), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 46/97 (47%), Gaps = 1/97 (1%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
I +R+DG D+ AT+ A P+LME TYR HS SD +YR E++
Sbjct 249 IPGLRVDGMDILCVREATRFAAAYCRSGKGPILMELQTYRYHGHSMSDPGVSYRTREEIQ 308
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRK 115
+ PI L+ + + + EE +E+ E RK
Sbjct 309 EVRSKSD-PIMLLKDRMVNSNLASVEELKEIDVEVRK 344
> Hs4885543
Length=388
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 46/97 (47%), Gaps = 1/97 (1%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
I +++DG D+ ATK A P+LME TYR HS SD +YR E++
Sbjct 247 IPGLKVDGMDVLCVREATKFAANYCRSGKGPILMELQTYRYHGHSMSDPGVSYRTREEIQ 306
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELRKESRK 115
S PI L+ + + T EE +E+ E RK
Sbjct 307 EVR-SKRDPIIILQDRMVNSKLATVEELKEIGAEVRK 342
> 7290525
Length=443
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 47/94 (50%), Gaps = 2/94 (2%)
Query 22 IRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELEAWN 81
I +DG D+ A AT+ A + +T+ P++ME TYR HS SD ++YR E++
Sbjct 300 IWVDGMDVLAVRSATEFAINYV-NTHGPLVMETNTYRYSGHSMSDPGTSYRTREEIQEVR 358
Query 82 ASGILPIARLRKYLAHEGWWTDEEEEELRKESRK 115
PI ++ G T +E + + + RK
Sbjct 359 QKRD-PITSFKELCIELGLITTDEVKAIDLKVRK 391
> ECU09g1040
Length=349
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 48/92 (52%), Gaps = 5/92 (5%)
Query 19 IQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELE 78
I IRI ++F K AR+ ++ N P++++ TYR HS +D+ +YR E++
Sbjct 232 IPGIRIGHGNIFGLMSVLKYARKYSVE-NGPIIVQIDTYRFCTHSAADERESYRSREEVD 290
Query 79 AWNASGILPIARLRKYLAHEGWWTDEEEEELR 110
A + R+ LA ++++EE + LR
Sbjct 291 AEKKRDCMEDVG-RRLLA---FYSEEELDALR 318
> 7290527
Length=479
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 46/95 (48%), Gaps = 8/95 (8%)
Query 24 IDGNDLFASFLATKQARQLILD---TNEPVLMEFMTYRVGQHSTSDDSSAYRPPGELEAW 80
+DGN + LA + A Q +D + P+++E TYR HS SD ++YR E+++
Sbjct 256 VDGNQV----LAVRSATQFAVDHALKHGPIVLEMSTYRYVGHSMSDPGTSYRSREEVQST 311
Query 81 NASGILPIARLRKYLAHEGWWTDEEEEELRKESRK 115
PI R + +EE + L ++RK
Sbjct 312 REKRD-PITSFRSQIIALCLADEEELKALDDKTRK 345
> At1g01090
Length=428
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Query 13 RGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAYR 72
+G ++G+ + +DG D+ K+A P L+E TYR HS +D R
Sbjct 276 KGPAFGMPGVHVDGMDVLKVREVAKEAVTRARRGEGPTLVECETYRFRGHSLADPDE-LR 334
Query 73 PPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKE 112
E + A PIA L+KYL + E + + K+
Sbjct 335 DAAEKAKYAARD--PIAALKKYLIENKLAKEAELKSIEKK 372
> At1g02980
Length=648
Score = 30.4 bits (67), Expect = 0.86, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 22 IRIDGNDLFASFLATKQARQLILDTN 47
+ I DLFA F KQAR+L+ D N
Sbjct 331 VYISDKDLFAEFFRKKQARRLLFDRN 356
> CE20160
Length=325
Score = 30.0 bits (66), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 2 RDQYAGDGIAIRGISYGIQTIRIDGNDLFASFLATKQARQLILDT 46
++ + + ++ ISY ++ I + F FL TK R L+L T
Sbjct 264 QEHFESLPLQVQSISYDLRLFAIGSLNFFVYFLVTKSIRNLVLRT 308
> 7297165
Length=992
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 24/56 (42%), Gaps = 7/56 (12%)
Query 59 VGQHSTSDDSSAYRPPGELEAWNASGILPIARLRKYLAHEGWWTDEEEEELRKESR 114
V HS ++ A PP S P+ Y+A W T E ELR++++
Sbjct 88 VPDHSNKENQPARTPP-------PSKSCPLGAPTNYVARRTWITTERMNELRRKAQ 136
> Hs22041150
Length=452
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 9/20 (45%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 8 DGIAIRGISYGIQTIRIDGN 27
+G+ ++G+ GIQ I+I+GN
Sbjct 53 NGVTLKGVKRGIQVIKIEGN 72
> ECU10g1210
Length=1799
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 90 RLRKYLAHEGWWTDEEEEELRKESRKY 116
R+RK + E + +E EELR+ +R+Y
Sbjct 611 RIRKQIRREKGFAGDENEELRQRARRY 637
> CE23525
Length=440
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 96 AHEGWWTDEEEEELRKESRKYMLEQ 120
AH+ WWT +E +++R E+ + ML
Sbjct 100 AHDLWWTGQEFKDMRVEACEQMLRH 124
> At1g43140
Length=711
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 22 IRIDGNDLFASFLATKQARQLILD 45
+ I DLFA F KQAR+L+ D
Sbjct 417 VYISDKDLFAEFYRKKQARRLLFD 440
> CE00494_2
Length=398
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 96 AHEGWWTDEEEEELRKESRKYMLEQ 120
AH+ WWT +E +++R E+ + ML
Sbjct 58 AHDLWWTGQEFKDMRVEACEQMLRH 82
> 7302338
Length=416
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 32/66 (48%), Gaps = 4/66 (6%)
Query 12 IRGISYGIQTIRIDGNDLFASFLATKQARQLILDTNEPVLMEFMTYRVGQHSTSDDSSAY 71
I+ + Y IQ I N ++A L A L + T+E ++ + +GQ + ++ S Y
Sbjct 239 IQELHYLIQEI----NRVYALSLWAAMAHDLAMSTSELYILFGQSVGIGQQNEEENGSCY 294
Query 72 RPPGEL 77
R G L
Sbjct 295 RMLGYL 300
Lambda K H
0.317 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40