bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3011_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
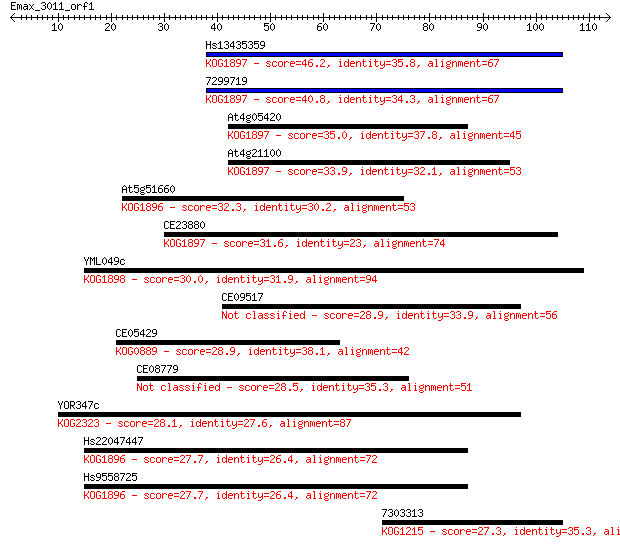
Hs13435359 46.2 2e-05
7299719 40.8 7e-04
At4g05420 35.0 0.035
At4g21100 33.9 0.073
At5g51660 32.3 0.22
CE23880 31.6 0.38
YML049c 30.0 1.1
CE09517 28.9 2.3
CE05429 28.9 2.6
CE08779 28.5 3.1
YOR347c 28.1 3.7
Hs22047447 27.7 5.0
Hs9558725 27.7 5.3
7303313 27.3 7.9
> Hs13435359
Length=1140
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 38 LDVLGDRYICVGDMMSSVCLVEWRPANQQLKEIARDMRCAWPLGVSVLDANSCIMSDGEQ 97
L GD +I VGD+M SV L+ ++P +EIARD W V +LD ++ + ++
Sbjct 914 LKTKGD-FILVGDLMRSVLLLAYKPMEGNFEEIARDFNPNWMSAVEILDDDNFLGAENAF 972
Query 98 QLWVLQR 104
L+V Q+
Sbjct 973 NLFVCQK 979
> 7299719
Length=1140
Score = 40.8 bits (94), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 38 LDVLGDRYICVGDMMSSVCLVEWRPANQQLKEIARDMRCAWPLGVSVLDANSCIMSDGEQ 97
L GD +I VGD+M S+ L++ + EIARD W V +LD ++ + S+
Sbjct 915 LKAKGD-FILVGDLMRSITLLQHKQMEGIFVEIARDCEPKWMRAVEILDDDTFLGSETNG 973
Query 98 QLWVLQR 104
L+V Q+
Sbjct 974 NLFVCQK 980
> At4g05420
Length=1088
Score = 35.0 bits (79), Expect = 0.035, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 42 GDRYICVGDMMSSVCLVEWRPANQQLKEIARDMRCAWPLGVSVLD 86
GD +I VGD+M S+ L+ ++ ++E ARD W V +LD
Sbjct 877 GD-FIVVGDLMKSISLLLYKHEEGAIEERARDYNANWMSAVEILD 920
> At4g21100
Length=1088
Score = 33.9 bits (76), Expect = 0.073, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 42 GDRYICVGDMMSSVCLVEWRPANQQLKEIARDMRCAWPLGVSVLDANSCIMSD 94
GD +I VGD+M S+ L+ ++ ++E ARD W V +L+ + + +D
Sbjct 877 GD-FIAVGDLMKSISLLIYKHEEGAIEERARDYNANWMTAVEILNDDIYLGTD 928
> At5g51660
Length=1448
Score = 32.3 bits (72), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 2/54 (3%)
Query 22 QLTPVAVFSAST-FIASLDVLGDRYICVGDMMSSVCLVEWRPANQQLKEIARDM 74
+L VA F A ++ S++V+ +I +GD+ S+ + W+ QL +A+D
Sbjct 1210 ELNGVAFFDAPPLYVVSMNVV-KSFILLGDVHKSIYFLSWKEQGSQLSLLAKDF 1262
> CE23880
Length=1134
Score = 31.6 bits (70), Expect = 0.38, Method: Composition-based stats.
Identities = 17/75 (22%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 30 SASTFIASLDV-LGDRYICVGDMMSSVCLVEWRPANQQLKEIARDMRCAWPLGVSVLDAN 88
S+ + +LD+ + + + V D+M SV L+ +R +E+A+D W + + A
Sbjct 899 SSFNHVIALDLKVMNEEVAVADVMRSVSLLSYRMLEGNFEEVAKDWNSQWMVTCEFITAE 958
Query 89 SCIMSDGEQQLWVLQ 103
S + + L+ ++
Sbjct 959 SILGGEAHLNLFTVE 973
> YML049c
Length=1361
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 30/101 (29%), Positives = 40/101 (39%), Gaps = 12/101 (11%)
Query 15 LSSLGRMQL-------TPVAVFSASTFIASLDVLGDRYICVGDMMSSVCLVEWRPANQQL 67
L LG+ QL TPV++ T I S+ + VGD+ SV L W PA
Sbjct 1051 LYGLGKKQLLRRSVTQTPVSI----TKIVSMHQWNYERLAVGDIHESVTLFIWDPAGNVF 1106
Query 68 KEIARDMRCAWPLGVSVLDANSCIMSDGEQQLWVLQRALPE 108
D + LD + I +D W L R+ PE
Sbjct 1107 IPYVDDSVKRHVTVLKFLDEATVIGADRYGNAWTL-RSPPE 1146
> CE09517
Length=290
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 6/59 (10%)
Query 41 LGDRYICVGDMMSSVCLVEWRPANQQLKEIAR---DMRCAWPLGVSVLDANSCIMSDGE 96
LG+R C +M C RP K AR + RC W +S A++C +S G+
Sbjct 49 LGNRCECPFGLMGRRC---QRPCQDVYKSCARWKSEERCHWTRPISPFFADNCALSCGQ 104
> CE05429
Length=3944
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 9/51 (17%)
Query 21 MQLTPVAVFSASTFIASL---------DVLGDRYICVGDMMSSVCLVEWRP 62
++++P++ +TF AS D DR+I D+M CLVE P
Sbjct 1491 LKVSPISTIIIATFGASYIRNISGAGDDSDSDRHISYNDIMKFKCLVELNP 1541
> CE08779
Length=715
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 7/58 (12%)
Query 25 PVAVFSASTFIASLD----VLGDRY---ICVGDMMSSVCLVEWRPANQQLKEIARDMR 75
P+A F FIA+ + G+R+ +C + + C PA +LKE RD+R
Sbjct 634 PLASFEVRRFIANDNRCAVCAGERHSSSMCYSTLKCTYCSGRHHPAGCRLKEFYRDVR 691
> YOR347c
Length=506
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 35/87 (40%), Gaps = 15/87 (17%)
Query 10 AAGDSLSSLGRMQLTPVAVFSASTFIASLDVLGDRYICVGDMMSSVCLVEWRPANQQLKE 69
A GD LG L P + IA ++ G IC M+ S+ RP ++ +
Sbjct 265 ARGD----LGIEILAPEVLAIQKKLIAKCNLAGKPVICATQMLDSMTH-NPRPTRAEVSD 319
Query 70 IARDMRCAWPLGVSVLDANSCIMSDGE 96
+ G +VLD C+M GE
Sbjct 320 V----------GNAVLDGADCVMLSGE 336
> Hs22047447
Length=1443
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 15 LSSLGRMQLTPVAVFSASTFIASLDVLGDRYICVGDMMSSVCLVEWRPANQQLKEIARDM 74
L SL +LT +A +I + + +I D+M S+ L+ ++ ++ L ++RD
Sbjct 1192 LWSLRASELTGMAFIDTQLYIHQM-ISVKNFILAADVMKSISLLRYQEESKTLSLVSRDA 1250
Query 75 RCAWPLGVSVLD 86
+ PL V +D
Sbjct 1251 K---PLEVYSVD 1259
> Hs9558725
Length=1442
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 15 LSSLGRMQLTPVAVFSASTFIASLDVLGDRYICVGDMMSSVCLVEWRPANQQLKEIARDM 74
L SL +LT +A +I + + +I D+M S+ L+ ++ ++ L ++RD
Sbjct 1192 LWSLRASELTGMAFIDTQLYIHQM-ISVKNFILAADVMKSISLLRYQEESKTLSLVSRDA 1250
Query 75 RCAWPLGVSVLD 86
+ PL V +D
Sbjct 1251 K---PLEVYSVD 1259
> 7303313
Length=1612
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 24/35 (68%), Gaps = 3/35 (8%)
Query 71 ARDMRCAWPLGVSVLDANSCIMSDGEQQ-LWVLQR 104
++ CA P GV ++ AN+C ++G Q+ ++++QR
Sbjct 312 SQGFSCACPTGVKLISANTC--ANGSQEMMFIVQR 344
Lambda K H
0.320 0.130 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1185472426
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40