bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3076_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
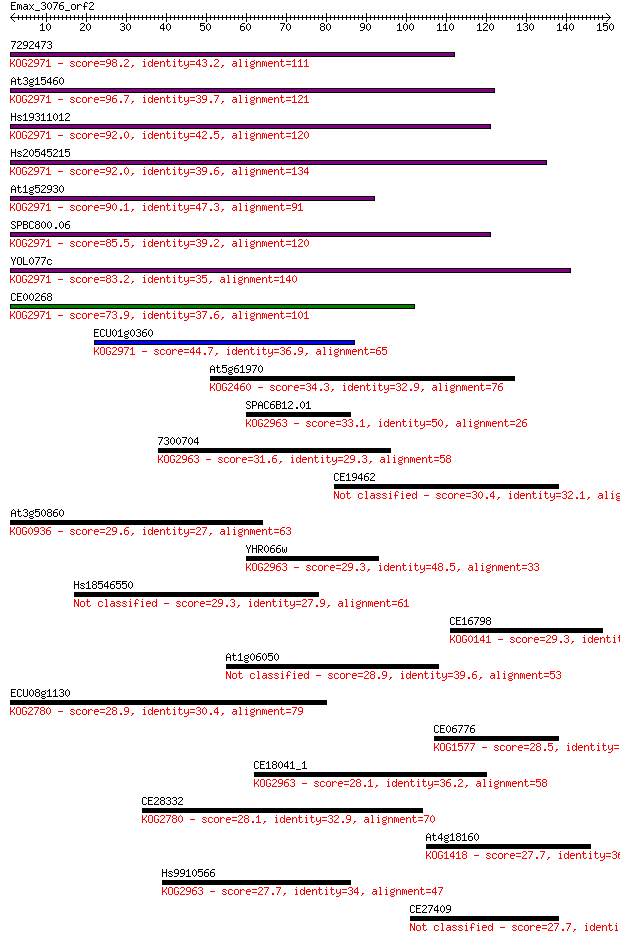
7292473 98.2 5e-21
At3g15460 96.7 2e-20
Hs19311012 92.0 4e-19
Hs20545215 92.0 4e-19
At1g52930 90.1 1e-18
SPBC800.06 85.5 4e-17
YOL077c 83.2 2e-16
CE00268 73.9 1e-13
ECU01g0360 44.7 7e-05
At5g61970 34.3 0.090
SPAC6B12.01 33.1 0.18
7300704 31.6 0.65
CE19462 30.4 1.2
At3g50860 29.6 2.0
YHR066w 29.3 2.7
Hs18546550 29.3 2.9
CE16798 29.3 2.9
At1g06050 28.9 3.4
ECU08g1130 28.9 3.8
CE06776 28.5 4.7
CE18041_1 28.1 6.7
CE28332 28.1 7.3
At4g18160 27.7 7.8
Hs9910566 27.7 8.0
CE27409 27.7 8.6
> 7292473
Length=356
Score = 98.2 bits (243), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 48/111 (43%), Positives = 68/111 (61%), Gaps = 18/111 (16%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KELF+Q + P +HPKS+PF DH +F D RIWFR++QI + +GG
Sbjct 171 KELFVQTYSVPNHHPKSQPFVDHVFTFTYLDKRIWFRNFQI---LSEDGG---------L 218
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKR 111
E+GPR+V++PVKI +GSF GKT+W N ++V P + R+VL A +
Sbjct 219 SEVGPRYVMNPVKIFDGSFTGKTIWENPDYV------SPSKQRQVLKKAAK 263
> At3g15460
Length=315
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/122 (39%), Positives = 69/122 (56%), Gaps = 1/122 (0%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIA-PLVMGEGGDANDPHRQT 59
KE+ Q+FG P H KSKP+ DH F + D IWFR+YQI+ P + D + T
Sbjct 176 KEMLTQIFGIPEGHRKSKPYHDHVFVFSIVDDHIWFRNYQISVPHNESDKIARGDLDKMT 235
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRR 119
IE+GPRF L+P+KI GSF G TL+ N +V + + + FAK+ K +R+
Sbjct 236 LIEVGPRFCLNPIKIFGGSFGGTTLYENPFYVSPNQIRALEKRNKAGKFAKKIKAKTRRK 295
Query 120 LY 121
++
Sbjct 296 MH 297
> Hs19311012
Length=354
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 51/129 (39%), Positives = 74/129 (57%), Gaps = 21/129 (16%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL +Q+F TPR HPKS+PF DH +F + D+RIWFR++QI DA
Sbjct 182 KELLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII------EEDA------AL 229
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEF---------VRTRDLMRPRRLREVLDFAKR 111
+EIGPRFVL+ +KI +GSF G TL+ N + +R+ + R ++V D ++
Sbjct 230 VEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPNMHRRVIRSITAAKYREKQQVKDVPQK 289
Query 112 QGQKEKRRL 120
+KE + L
Sbjct 290 LRKKEPKTL 298
> Hs20545215
Length=353
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 53/134 (39%), Positives = 76/134 (56%), Gaps = 17/134 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL +Q+F TPR HPKS+PF DH +F + D+RIWFR++QI DA
Sbjct 182 KELLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII------EEDA------AL 229
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRL 120
+EIGPRFVL+ +KI +GSF G TL+ N + M R +R + R+ Q+ K
Sbjct 230 VEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPN--MHRRVIRSITAAKYREKQQVKD-- 285
Query 121 YLESMMEGEPRNKL 134
++ + + EP+ L
Sbjct 286 -VQKLRKKEPKTLL 298
> At1g52930
Length=320
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIA-PLVMGEGGDANDPHRQT 59
KE+ QVFG P+ H KSKP+ DH F + D IWFR+YQI+ P + + T
Sbjct 180 KEMLTQVFGIPKEHRKSKPYHDHVFVFSIVDEHIWFRNYQISVPHNESDKIAKGGLDKMT 239
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFV 91
IE+GPRF L+P+KI GSF G TL+ N +V
Sbjct 240 LIEVGPRFCLNPIKIFAGSFGGPTLYENPLYV 271
> SPBC800.06
Length=295
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/121 (38%), Positives = 67/121 (55%), Gaps = 8/121 (6%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGD-ANDPHRQT 59
KEL Q FG P+ +SKPF D + + D +IWFR+Y+I E D + DP T
Sbjct 165 KELLQQTFGIPKGARRSKPFIDRVCTLTIADGKIWFRNYEIR-----ENEDKSKDP--VT 217
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRR 119
IEIGPRFV+ + ILEGSF G +++N FV + + R + + RQ K +R+
Sbjct 218 LIEIGPRFVMTIINILEGSFGGPVIYKNDTFVSSTMVRAAIRNQAAQRYVNRQESKLERQ 277
Query 120 L 120
+
Sbjct 278 V 278
> YOL077c
Length=291
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 49/145 (33%), Positives = 81/145 (55%), Gaps = 12/145 (8%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL + F P N KSKPF DH MSF + D +IW R Y+I+ + + +
Sbjct 153 KELLVHNFCVPPNARKSKPFIDHVMSFSIVDDKIWVRTYEISHSTKNKEEYEDGEEDISL 212
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQ-----K 115
+EIGPRFV+ + ILEGSF G ++ N ++V + +++R + ++ + AK + + K
Sbjct 213 VEIGPRFVMTVILILEGSFGGPKIYENKQYV-SPNVVRAQIKQQAAEEAKSRAEAAVERK 271
Query 116 EKRRLYLESMMEGEPRNKLAKEVVF 140
KRR E+++ +P L+ + +F
Sbjct 272 IKRR---ENVLAADP---LSNDALF 290
> CE00268
Length=352
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 55/102 (53%), Gaps = 13/102 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTD-SRIWFRHYQIAPLVMGEGGDANDPHRQT 59
K + MQ GTP +HP+S+PF DH +F V + +IWFR++QI +
Sbjct 185 KAVLMQTLGTPHHHPRSQPFVDHVFNFSVGEGDKIWFRNFQIVDESL------------Q 232
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRR 101
E+GPRFVL+ V++ GSF G L+ N +V + R R
Sbjct 233 LQEVGPRFVLEMVRLFAGSFEGAVLYDNPNYVSPNVIRREHR 274
> ECU01g0360
Length=196
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 32/65 (49%), Gaps = 15/65 (23%)
Query 22 DHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILEGSFCG 81
D A+ FF D IW R Y+I + EIGPR VL+ +K+LE F G
Sbjct 134 DKALCFFYLDGVIWVRCYKIG---------------KKLEEIGPRLVLEVLKVLEKCFEG 178
Query 82 KTLWR 86
K L++
Sbjct 179 KALYK 183
> At5g61970
Length=627
Score = 34.3 bits (77), Expect = 0.090, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 43/78 (55%), Gaps = 4/78 (5%)
Query 51 DANDPHRQTFIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDL--MRPRRLREVLDF 108
+ +DP ++ +I PRF ++ +++++ S L R+G++ R R R RRL + L F
Sbjct 12 EIDDPKSESSDQILPRFSINVLQLMKSSQAQHGL-RHGDYARYRRYCSARLRRLYKSLKF 70
Query 109 AKRQGQKEKRRLYLESMM 126
+G K RR LES +
Sbjct 71 THGRG-KYTRRAILESTV 87
> SPAC6B12.01
Length=361
Score = 33.1 bits (74), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLW 85
IEIGPR L+ +KI E + GK L+
Sbjct 261 LIEIGPRMTLELIKITEDAMGGKVLY 286
> 7300704
Length=460
Score = 31.6 bits (70), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 38 HYQIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRD 95
H +A + +G + EIGPR + +KI EG G+ L+ + V+T D
Sbjct 248 HVVLAQTLKSKGNLEDKKSSIKLHEIGPRLTMQLIKIEEGLLTGEVLYHD-HVVKTED 304
> CE19462
Length=149
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 82 KTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRLYLESMMEGE-PRNKLAKE 137
K ++N EF+R + + R + +++AK+Q +K KR++ L S + E NK A +
Sbjct 7 KFFFKNSEFIR--NFLEIRVIIRNMEYAKKQAEKHKRKIILPSYISWEYGSNKFASQ 61
> At3g50860
Length=145
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 28/63 (44%), Gaps = 0/63 (0%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
+EL VF + P++ F S F DSR+ ++HY V+ G N+
Sbjct 29 QELIRGVFSVLCSRPENVSNFLEIESLFGPDSRLVYKHYATLYFVLVFDGSENELAMLDL 88
Query 61 IEI 63
I++
Sbjct 89 IQV 91
> YHR066w
Length=453
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVR 92
E+GPR L VKI EG GK L EFV+
Sbjct 328 LTELGPRLTLKLVKIEEGICSGKVLHH--EFVQ 358
> Hs18546550
Length=362
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 25/61 (40%), Gaps = 2/61 (3%)
Query 17 SKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILE 76
+KP F + + W RH I + GG+ D +T P F LDP ++
Sbjct 278 TKPHFASLARMRSSRWQDWVRHDTIVCIEEARGGERTDSQSKT--SSTPEFCLDPTRVAT 335
Query 77 G 77
G
Sbjct 336 G 336
> CE16798
Length=419
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 111 RQGQKEKRRLYLESMMEGEPRNKLA-KEVVFGSDVVKQQ 148
R G +E+R+ YL ++ GE LA E GSDVV +
Sbjct 136 RNGSEEQRKKYLPKLISGEHMGALAMSEAQAGSDVVSMK 174
> At1g06050
Length=313
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 28/57 (49%), Gaps = 4/57 (7%)
Query 55 PHRQTFIEIGPRFVLDPVKILEGSFCGKTL---WRNGEFVRTRDLMRP-RRLREVLD 107
P TF+ GP++ D VKI G F K L W G + L P R+R+V+D
Sbjct 39 PSPDTFMVRGPKYFSDKVKIPAGDFLLKPLGFDWIKGPKKLSEILSYPSSRIRKVID 95
> ECU08g1130
Length=177
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 34/79 (43%), Gaps = 14/79 (17%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KELF+ + P+ F + F+ I RHY I + E D + F
Sbjct 112 KELFLSLSSRPKT-------FKRNLHFYFDGDLIHVRHYAI---LTKEEEDI----KVGF 157
Query 61 IEIGPRFVLDPVKILEGSF 79
EIGPR + VK ++G F
Sbjct 158 HEIGPRITMRLVKKMDGFF 176
> CE06776
Length=317
Score = 28.5 bits (62), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 107 DFAKRQGQKEKRRLYLESMMEGEPRNKLAKE 137
D AK + K +RL+L+ M G P + A E
Sbjct 285 DIAKLEESKNSQRLFLQDFMTGHPEDAFAAE 315
> CE18041_1
Length=400
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 31/58 (53%), Gaps = 6/58 (10%)
Query 62 EIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRR 119
EIGPR L+ VKI EG G+ L+ +L+ +LR +D ++ Q +KRR
Sbjct 342 EIGPRLTLELVKIEEGIDEGEVLYHKHNAKTPDELI---KLRAHMD---KKKQMKKRR 393
> CE28332
Length=384
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 32/70 (45%), Gaps = 10/70 (14%)
Query 34 IWFRHYQIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRT 93
I+FRH++ EG A +E+GPRF L + +G+F K W E+V
Sbjct 322 IFFRHHRYE--FKKEGSKA------ALLELGPRFTLRLKWLQKGTFDAK--WGEFEWVLK 371
Query 94 RDLMRPRRLR 103
R M R R
Sbjct 372 RHEMETSRRR 381
> At4g18160
Length=436
Score = 27.7 bits (60), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 105 VLDFAKRQGQKEKRRLYLESMMEGEPRNKLAKEVVFGSDVV 145
+LD AKR+ + EKRR Y+ + +G R +L + G V+
Sbjct 243 MLDSAKRRDEPEKRRSYIIDVKKGRMRIRLKVALALGVVVL 283
> Hs9910566
Length=473
Score = 27.7 bits (60), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query 39 YQIAPLVMGEGGDANDPHRQTFI---EIGPRFVLDPVKILEGSFCGKTLW 85
+ I L G N +Q+ + EIGPR L +K+ EG GK ++
Sbjct 248 HNITELPQAVAGRGNMRAQQSAVRLTEIGPRMTLQLIKVQEGVGEGKVMF 297
> CE27409
Length=609
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 101 RLREVLDFAKRQGQKEKRRLYLESMMEGEPRNKLAKE 137
R R+++ F Q+E+R+ MEG+ NK K+
Sbjct 93 RARDIVAFTSYMAQEEERKRKTREQMEGDNSNKTPKK 129
Lambda K H
0.325 0.141 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40