bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3147_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
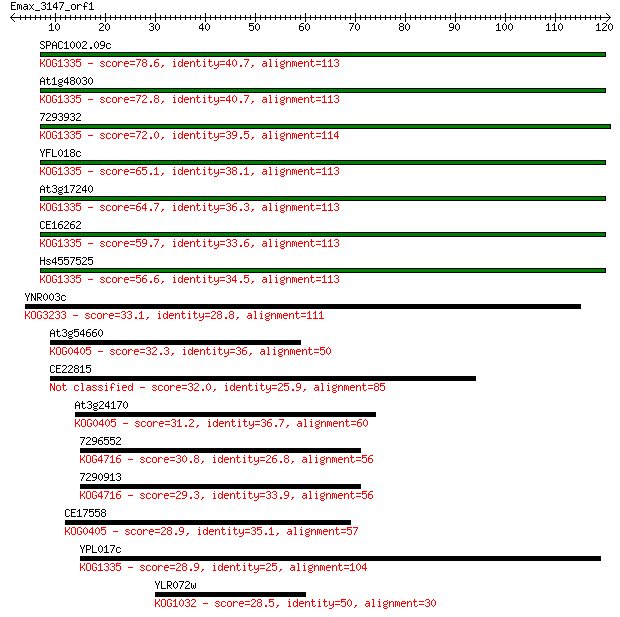
SPAC1002.09c 78.6 3e-15
At1g48030 72.8 1e-13
7293932 72.0 3e-13
YFL018c 65.1 3e-11
At3g17240 64.7 3e-11
CE16262 59.7 1e-09
Hs4557525 56.6 1e-08
YNR003c 33.1 0.11
At3g54660 32.3 0.23
CE22815 32.0 0.31
At3g24170 31.2 0.47
7296552 30.8 0.55
7290913 29.3 1.7
CE17558 28.9 2.2
YPL017c 28.9 2.6
YLR072w 28.5 3.0
> SPAC1002.09c
Length=511
Score = 78.6 bits (192), Expect = 3e-15, Method: Composition-based stats.
Identities = 46/113 (40%), Positives = 58/113 (51%), Gaps = 31/113 (27%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GIE LFK+NKVEY KG G DP T+ V+ I G + + +K KN I+ATGSE P G
Sbjct 137 LTSGIEYLFKKNKVEYAKGTGSFIDPQTLSVKGIDGAADQTIKAKNFIIATGSEVKPFPG 196
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE++IVSSTGAL+LS VP+
Sbjct 197 -------------------------------VTIDEKKIVSSTGALSLSEVPK 218
> At1g48030
Length=507
Score = 72.8 bits (177), Expect = 1e-13, Method: Composition-based stats.
Identities = 46/113 (40%), Positives = 62/113 (54%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L +GIEGLFK+NKV Y+KG G+ P+ V V+ I GG+T +K K+II+ATGS+ L G
Sbjct 135 LTRGIEGLFKKNKVTYVKGYGKFISPNEVSVETIDGGNT-IVKGKHIIVATGSDVKSLPG 193
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE++IVSSTGAL+LS VP+
Sbjct 194 -------------------------------ITIDEKKIVSSTGALSLSEVPK 215
> 7293932
Length=504
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/114 (39%), Positives = 59/114 (51%), Gaps = 32/114 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV + G G + +P+ VEV+ GSTE +KTKNI++ATGSE P G
Sbjct 130 LTGGIAMLFKKNKVTQLTGFGTIVNPNEVEVKK-SDGSTETVKTKNILIATGSEVTPFPG 188
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPRH 120
++DE+ IVSSTGAL L+ VP+H
Sbjct 189 -------------------------------IEIDEEVIVSSTGALKLAKVPKH 211
> YFL018c
Length=499
Score = 65.1 bits (157), Expect = 3e-11, Method: Composition-based stats.
Identities = 43/118 (36%), Positives = 57/118 (48%), Gaps = 36/118 (30%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGG--GSTEK---LKTKNIILATGSEA 61
L GIE LFK+NKV Y KG G D + V P+ G G+ ++ L KNII+ATGSE
Sbjct 119 LTGGIELLFKKNKVTYYKGNGSFEDETKIRVTPVDGLEGTVKEDHILDVKNIIVATGSEV 178
Query 62 APLVGGALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
P G ++DE++IVSSTGAL+L +P+
Sbjct 179 TPFPG-------------------------------IEIDEEKIVSSTGALSLKEIPK 205
> At3g17240
Length=507
Score = 64.7 bits (156), Expect = 3e-11, Method: Composition-based stats.
Identities = 41/113 (36%), Positives = 58/113 (51%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L +G+EGLFK+NKV Y+KG G+ P V V I G + +K K+II+ATGS+ L G
Sbjct 135 LTRGVEGLFKKNKVNYVKGYGKFLSPSEVSVDTIDGENV-VVKGKHIIVATGSDVKSLPG 193
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE++IVSSTGAL+L+ +P+
Sbjct 194 -------------------------------ITIDEKKIVSSTGALSLTEIPK 215
> CE16262
Length=464
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 55/113 (48%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI+ LFK NKV +++G + P+TV+ + GS E + +NI++A+GSE P G
Sbjct 120 LTGGIKQLFKANKVGHVEGFATIVGPNTVQAKK-NDGSVETINARNILIASGSEVTPFPG 178
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE+QIVSSTGAL+L VP+
Sbjct 179 -------------------------------ITIDEKQIVSSTGALSLGQVPK 200
> Hs4557525
Length=509
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 51/113 (45%), Gaps = 32/113 (28%)
Query 7 LAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVG 66
L GI LFK+NKV ++ G G++ + V GG T+ + TKNI++ATGSE P G
Sbjct 134 LTGGIAHLFKQNKVVHVNGYGKITGKNQVTATKADGG-TQVIDTKNILIATGSEVTPFPG 192
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVPR 119
+DE IVSSTGAL+L VP
Sbjct 193 -------------------------------ITIDEDTIVSSTGALSLKKVPE 214
> YNR003c
Length=317
Score = 33.1 bits (74), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 57/124 (45%), Gaps = 14/124 (11%)
Query 4 SCVLAQGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIIL-------- 55
S ++++GI LF + +++ G G L D ++ VQ + + KL +N L
Sbjct 20 SQMMSKGIGALFTQQELQKQMGIGSLTDLMSI-VQELLDKNLIKLVKQNDELKFQGVLES 78
Query 56 -----ATGSEAAPLVGGALEAVTCSRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTG 110
AT S LV +EA + +K + +RT+ Q +V CL ++ Q+ V S
Sbjct 79 EAQKKATMSAEEALVYSYIEASGREGIWSKTIKARTNLHQHVVLKCLKSLESQRYVKSVK 138
Query 111 ALAL 114
++
Sbjct 139 SVKF 142
> At3g54660
Length=565
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 10/54 (18%)
Query 9 QGIEGLFK----RNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATG 58
Q + G++K + V+ I+G G++ DPHTV+V + T+NI++A G
Sbjct 186 QRLTGIYKNILSKANVKLIEGRGKVIDPHTVDVD------GKIYTTRNILIAVG 233
> CE22815
Length=355
Score = 32.0 bits (71), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 42/87 (48%), Gaps = 2/87 (2%)
Query 9 QGIEGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGST--EKLKTKNIILATGSEAAPLVG 66
+ ++ + +K Y +G+G P +V I G ++ E+ TKN +T +P++
Sbjct 176 KSLQTDYSDDKFLYSEGSGLTKSPIDYDVYQIMGSASDIEESPTKNSFHSTLENLSPILK 235
Query 67 GALEAVTCSRLSTKHLSSRTSRFQRLV 93
+ + S +ST+ SS + F LV
Sbjct 236 HDMVVASSSEISTQSQSSLKAVFHDLV 262
> At3g24170
Length=499
Score = 31.2 bits (69), Expect = 0.47, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query 14 LFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEA-APLVGGALEAV 72
L V+ +G GR+ P+ VEV+ I G K+I++ATGS A P + G A+
Sbjct 133 LLANAAVKLYEGEGRVVGPNEVEVRQIDGTKIS-YTAKHILIATGSRAQKPNIPGHELAI 191
Query 73 T 73
T
Sbjct 192 T 192
> 7296552
Length=516
Score = 30.8 bits (68), Expect = 0.55, Method: Composition-based stats.
Identities = 15/57 (26%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 15 FKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAA-PLVGGALE 70
+ KVEY+ D HT+E + G ++ ++ +++A G P + GA+E
Sbjct 141 LRDKKVEYVNSMATFRDSHTIEYVAMPGAEHRQVTSEYVVVAVGGRPRYPDIPGAVE 197
> 7290913
Length=491
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 26/56 (46%), Gaps = 1/56 (1%)
Query 15 FKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVGGALE 70
+ KVEYI G G D HT+ + G T +T +I G P + GA+E
Sbjct 118 LRDKKVEYINGLGSFVDSHTLLAKLKSGERTITAQT-FVIAVGGRPRYPDIPGAVE 172
> CE17558
Length=473
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 12 EGLFKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAA-PLVGGA 68
E K + VEYI+G A+ TVEV + K + KN ++A G + P + GA
Sbjct 115 ESGLKGSSVEYIRGRATFAEDGTVEV------NGAKYRGKNTLIAVGGKPTIPNIKGA 166
> YPL017c
Length=499
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 37/104 (35%), Gaps = 32/104 (30%)
Query 15 FKRNKVEYIKGAGRLADPHTVEVQPIGGGSTEKLKTKNIILATGSEAAPLVGGALEAVTC 74
+N V KG DPH VE+ G ++ K I++ATGS G A
Sbjct 119 LSKNNVTVYKGTAAFKDPHHVEIAQ-RGMKPFIVEAKYIVVATGSAVIQCPGVA------ 171
Query 75 SRLSTKHLSSRTSRFQRLVFFCLCQVDEQQIVSSTGALALSSVP 118
+D +I+SS AL+L +P
Sbjct 172 -------------------------IDNDKIISSDKALSLDYIP 190
> YLR072w
Length=693
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 18/35 (51%), Gaps = 5/35 (14%)
Query 30 ADPHTVEVQPI-----GGGSTEKLKTKNIILATGS 59
ADPH +V I GGG T NII+AT +
Sbjct 69 ADPHLSDVNSILDNHRGGGETALTSVNNIIMATST 103
Lambda K H
0.317 0.132 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40