bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
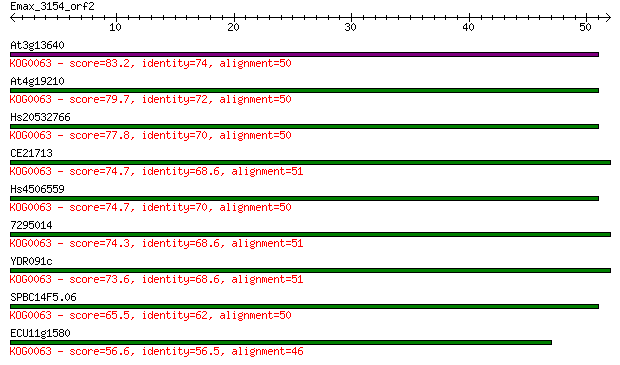
Query= Emax_3154_orf2
Length=51
Score E
Sequences producing significant alignments: (Bits) Value
At3g13640 83.2 1e-16
At4g19210 79.7 1e-15
Hs20532766 77.8 4e-15
CE21713 74.7 4e-14
Hs4506559 74.7 4e-14
7295014 74.3 5e-14
YDR091c 73.6 9e-14
SPBC14F5.06 65.5 2e-11
ECU11g1580 56.6 1e-08
> At3g13640
Length=603
Score = 83.2 bits (204), Expect = 1e-16, Method: Composition-based stats.
Identities = 37/50 (74%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
SL++GMN FL LNITFRRDPTNFRPRINKL+S KDKEQK G+++ LDD
Sbjct 554 SLLSGMNHFLSHLNITFRRDPTNFRPRINKLESIKDKEQKTAGSYYYLDD 603
> At4g19210
Length=600
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 36/50 (72%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
SL++GMN FL LNITFRRDPTNFRPRINKL+S KD+EQK G+++ LDD
Sbjct 551 SLLSGMNLFLSHLNITFRRDPTNFRPRINKLESTKDREQKSAGSYYYLDD 600
> Hs20532766
Length=599
Score = 77.8 bits (190), Expect = 4e-15, Method: Composition-based stats.
Identities = 35/50 (70%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP N+RPRINKL+S KD EQK +GN+F LDD
Sbjct 550 TLLAGMNKFLSQLEITFRRDPNNYRPRINKLNSIKDVEQKKSGNYFFLDD 599
> CE21713
Length=610
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/51 (68%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDDN 51
SL+ GMN+FLK+L+ITFRRD +RPRINKLDS KD +QK +G FF LDDN
Sbjct 560 SLLEGMNRFLKMLDITFRRDQETYRPRINKLDSVKDVDQKKSGQFFFLDDN 610
> Hs4506559
Length=599
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/50 (70%), Positives = 41/50 (82%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
+L+ GMNKFL L ITFRRDP N+RPRINKL+S KD EQK +GN+F LDD
Sbjct 550 TLLAGMNKFLSQLEITFRRDPNNYRPRINKLNSIKDVEQKKSGNYFFLDD 599
> 7295014
Length=611
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/51 (68%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDDN 51
SL+NGMN+FL++L ITFRRDP NFRPRINK +S KD EQK +G FF L+D
Sbjct 558 SLLNGMNRFLELLGITFRRDPNNFRPRINKNNSVKDTEQKRSGQFFFLEDE 608
> YDR091c
Length=608
Score = 73.6 bits (179), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 35/51 (68%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDDN 51
SL+ G N+FLK LN+TFRRDP +FRPRINKLDS DKEQK +GN+F LD+
Sbjct 556 SLLTGCNRFLKNLNVTFRRDPNSFRPRINKLDSQMDKEQKSSGNYFFLDNT 606
> SPBC14F5.06
Length=593
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/50 (62%), Positives = 38/50 (76%), Gaps = 0/50 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFFVLDD 50
SL+ GMN FLK L++TFRRDP RPRINK DS D+EQK GN+F L++
Sbjct 544 SLLTGMNTFLKNLDVTFRRDPNTLRPRINKFDSQMDQEQKNAGNYFFLEN 593
> ECU11g1580
Length=624
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/46 (56%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 1 SLINGMNKFLKVLNITFRRDPTNFRPRINKLDSCKDKEQKLNGNFF 46
L++GMN FLK L++TFRRD +N RPR+NK S KD+ QK N +F
Sbjct 577 GLLDGMNIFLKSLDVTFRRDSSNLRPRVNKPGSAKDRIQKENNQYF 622
Lambda K H
0.323 0.142 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161614636
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40