bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3174_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
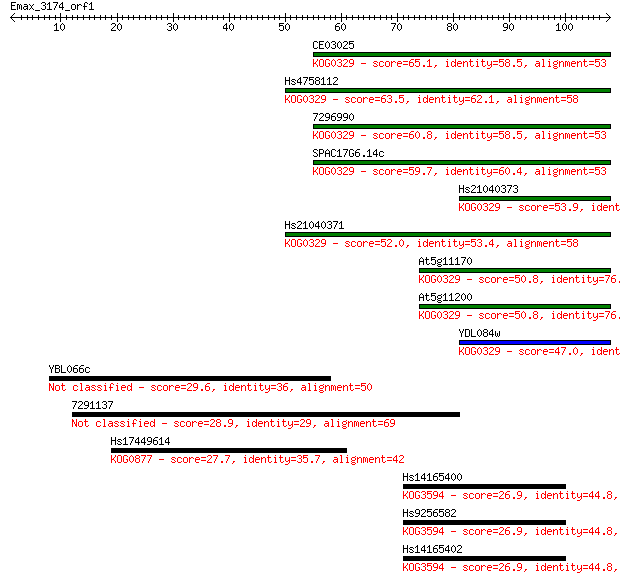
CE03025 65.1 3e-11
Hs4758112 63.5 8e-11
7296990 60.8 6e-10
SPAC17G6.14c 59.7 1e-09
Hs21040373 53.9 7e-08
Hs21040371 52.0 3e-07
At5g11170 50.8 5e-07
At5g11200 50.8 5e-07
YDL084w 47.0 8e-06
YBL066c 29.6 1.4
7291137 28.9 2.2
Hs17449614 27.7 5.1
Hs14165400 26.9 8.8
Hs9256582 26.9 8.9
Hs14165402 26.9 9.4
> CE03025
Length=425
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 42/55 (76%), Gaps = 2/55 (3%)
Query 55 DELVDYEEDEQ--QNDTKEKGVGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
++L+DYEE+++ Q+ E G G+ +G Y SIH+SGFRDFLLKPE+LRAIGD
Sbjct 4 EQLLDYEEEQEEIQDKQPEVGGGDARKTKGTYASIHSSGFRDFLLKPEILRAIGD 58
> Hs4758112
Length=428
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 36/62 (58%), Positives = 44/62 (70%), Gaps = 6/62 (9%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVGR----GNYVSIHASGFRDFLLKPELLRAI 105
E + +EL+DYE+DE + T G G EA + G+YVSIH+SGFRDFLLKPELLRAI
Sbjct 3 ENDVDNELLDYEDDEVE--TAAGGDGAEAPAKKDVKGSYVSIHSSGFRDFLLKPELLRAI 60
Query 106 GD 107
D
Sbjct 61 VD 62
> 7296990
Length=424
Score = 60.8 bits (146), Expect = 6e-10, Method: Composition-based stats.
Identities = 31/54 (57%), Positives = 39/54 (72%), Gaps = 1/54 (1%)
Query 55 DELVDYEEDEQQNDTK-EKGVGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
D+L+DYE++EQ T E + +G YVSIH+SGFRDFLLKPE+LRAI D
Sbjct 5 DDLLDYEDEEQTETTAVENQEAPKKDVKGTYVSIHSSGFRDFLLKPEILRAIVD 58
> SPAC17G6.14c
Length=434
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 43/65 (66%), Gaps = 14/65 (21%)
Query 55 DELVDYEEDEQ------------QNDTKEKGVGEEAVGRGNYVSIHASGFRDFLLKPELL 102
++L+DYEE+E+ DT E G E++ +G+YV IH++GFRDFLLKPELL
Sbjct 6 EDLIDYEEEEELVQDQPAQEITPAADTAENG--EKSDKKGSYVGIHSTGFRDFLLKPELL 63
Query 103 RAIGD 107
RAI D
Sbjct 64 RAITD 68
> Hs21040373
Length=316
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 23/27 (85%), Positives = 26/27 (96%), Gaps = 0/27 (0%)
Query 81 RGNYVSIHASGFRDFLLKPELLRAIGD 107
+G+YVSIH+SGFRDFLLKPELLRAI D
Sbjct 35 KGSYVSIHSSGFRDFLLKPELLRAIVD 61
> Hs21040371
Length=427
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 31/59 (52%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVG-RGNYVSIHASGFRDFLLKPELLRAIGD 107
EQ+ ++L+DY+E+E+ +E +G+YVSIH+SGFRDFLLKPELLRAI D
Sbjct 3 EQDVENDLLDYDEEEEPQAPQESTPAPPKKDIKGSYVSIHSSGFRDFLLKPELLRAIVD 61
> At5g11170
Length=435
Score = 50.8 bits (120), Expect = 5e-07, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 28/34 (82%), Gaps = 1/34 (2%)
Query 74 VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
V EAV +G YV IH+SGFRDFLLKPELLRAI D
Sbjct 31 VNGEAVKKG-YVGIHSSGFRDFLLKPELLRAIVD 63
> At5g11200
Length=427
Score = 50.8 bits (120), Expect = 5e-07, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 28/34 (82%), Gaps = 1/34 (2%)
Query 74 VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
V EAV +G YV IH+SGFRDFLLKPELLRAI D
Sbjct 31 VNGEAVKKG-YVGIHSSGFRDFLLKPELLRAIVD 63
> YDL084w
Length=446
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 19/27 (70%), Positives = 24/27 (88%), Gaps = 0/27 (0%)
Query 81 RGNYVSIHASGFRDFLLKPELLRAIGD 107
+G+YV IH++GF+DFLLKPEL RAI D
Sbjct 53 KGSYVGIHSTGFKDFLLKPELSRAIID 79
> YBL066c
Length=1057
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 8 NRCETTFALLASSLAQEAAATAATAAVAPPPQTPPLFKMAALEQNPADEL 57
N + L ASSLAQ++ AT P + PPL +AL N + L
Sbjct 205 NEASSHMTLRASSLAQDSKGLVATE----PNKLPPLLNDSALPNNSKESL 250
> 7291137
Length=109
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 9/71 (12%)
Query 12 TTFALLASS--LAQEAAATAATAAVAPPPQTPPLFKMAALEQNPADELVDYEEDEQQNDT 69
+TF++ +S+ L +A ++ +V PP ++ N + L+DYE+ Q
Sbjct 41 STFSMRSSAGELTSTSAPLSSIVSVTAPP-----MPFGGVKYNQSSGLIDYEKRNQV--V 93
Query 70 KEKGVGEEAVG 80
K KG GE + G
Sbjct 94 KVKGGGERSCG 104
> Hs17449614
Length=194
Score = 27.7 bits (60), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 19 SSLAQEAAATAATAAVAPPPQTPPLFKMAALEQNPADELVDY 60
S+L A AT T + QTP L+K ++P E DY
Sbjct 123 SALGNFAKATFDTISKTYSYQTPALWKETVFTKSPYQEFTDY 164
> Hs14165400
Length=807
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 2/29 (6%)
Query 71 EKGVGEEAVGRGNYVSIHASGFRDFLLKP 99
E+ VGE A+ NYVS+HA + + L+P
Sbjct 496 ERRVGERALS--NYVSVHAESGKVYALQP 522
> Hs9256582
Length=950
Score = 26.9 bits (58), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 2/29 (6%)
Query 71 EKGVGEEAVGRGNYVSIHASGFRDFLLKP 99
E+ VGE A+ NYVS+HA + + L+P
Sbjct 496 ERRVGERALS--NYVSVHAESGKVYALQP 522
> Hs14165402
Length=686
Score = 26.9 bits (58), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 2/29 (6%)
Query 71 EKGVGEEAVGRGNYVSIHASGFRDFLLKP 99
E+ VGE A+ NYVS+HA + + L+P
Sbjct 496 ERRVGERALS--NYVSVHAESGKVYALQP 522
Lambda K H
0.311 0.128 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40