bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
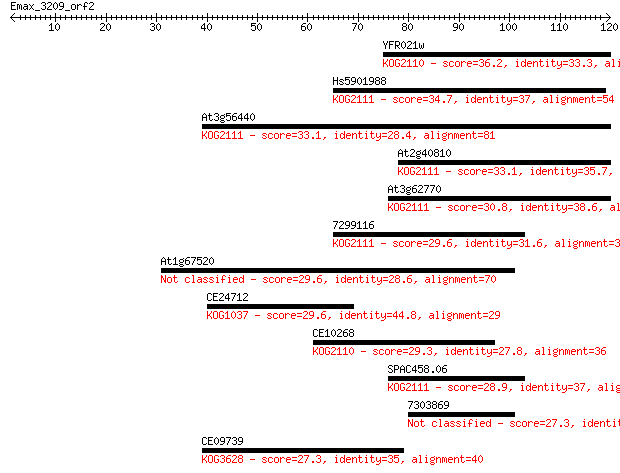
Query= Emax_3209_orf2
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
YFR021w 36.2 0.014
Hs5901988 34.7 0.038
At3g56440 33.1 0.13
At2g40810 33.1 0.13
At3g62770 30.8 0.71
7299116 29.6 1.3
At1g67520 29.6 1.4
CE24712 29.6 1.4
CE10268 29.3 1.8
SPAC458.06 28.9 2.5
7303869 27.3 6.7
CE09739 27.3 7.2
> YFR021w
Length=500
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 75 FLGFNSDASCLCVGTTAGFLVWSVRPLKCMYTCVCGPVTLVEMLL 119
F+ FN +C+ +GT+ GF +++ P Y+ G +VEML
Sbjct 10 FINFNQTGTCISLGTSKGFKIFNCEPFGKFYSEDSGGYAIVEMLF 54
> Hs5901988
Length=360
Score = 34.7 bits (78), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query 65 SSQPVKAATTFLGFNSDASCLCVGTTAGFLVWSVRPLK---CMYTCVCGPVTLVEML 118
+ QP++ T+ L FN D SC C G +++V PL + G + LVEML
Sbjct 2 TQQPLRGVTS-LRFNQDQSCFCCAMETGVRIYNVEPLMEKGHLDHEQVGSMGLVEML 57
> At3g56440
Length=400
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 36/83 (43%), Gaps = 4/83 (4%)
Query 39 RRSSIGGISSSKTMSTAAAEAGLSSGSSQPVKAATTFLGFNSDASCLCVGTTAGFLVWSV 98
RR+ G S+ T+ + G S +A + +N D SC GT+ GF +++
Sbjct 4 RRNFQPGGYDSRNTFTSGSFGPPDFGESD--EAELVSVSWNQDYSCFAAGTSHGFRIYNC 61
Query 99 RPLKCMY--TCVCGPVTLVEMLL 119
P K + G +VEML
Sbjct 62 EPFKETFRRELKDGGFKIVEMLF 84
> At2g40810
Length=369
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 78 FNSDASCLCVGTTAGFLVWSVRPLKCMY--TCVCGPVTLVEMLL 119
+N D+SC GT+ GF +++ P K + G +VEML
Sbjct 13 WNQDSSCFAAGTSHGFRIYNCEPFKETFRRELKDGGFKIVEMLF 56
> At3g62770
Length=432
Score = 30.8 bits (68), Expect = 0.71, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 3/47 (6%)
Query 76 LGFNSDASCLCVGTTAGFLVWSVRPLKCMYTC---VCGPVTLVEMLL 119
L FN D +C VGT GF + + P + ++ G V +VEML
Sbjct 81 LSFNQDHACFAVGTDRGFRILNCDPFREIFRRDFDRGGGVAVVEMLF 127
> 7299116
Length=340
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 15/38 (39%), Gaps = 0/38 (0%)
Query 65 SSQPVKAATTFLGFNSDASCLCVGTTAGFLVWSVRPLK 102
P + FN D C T GF V++ PLK
Sbjct 5 EQNPYGNGLLYAAFNQDQGCFACATDTGFRVYNCDPLK 42
> At1g67520
Length=743
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 34/74 (45%), Gaps = 7/74 (9%)
Query 31 CIGHLYIVRRSSIGGISSSKTMSTAAAEAGLSSGSSQPVKAATTFLGFNSDASCLCVGTT 90
C+ Y+VR G S T+S++A+ + SG+ V + L ++SCL +T
Sbjct 290 CLAAGYVVRDEPYGFTSFRVTVSSSASNGFVLSGTFSSVDCSAICL---QNSSCLAYAST 346
Query 91 ----AGFLVWSVRP 100
G +W+ P
Sbjct 347 EPDGTGCEIWNTYP 360
> CE24712
Length=2276
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 40 RSSIGGISSSKTMSTAAAEAGLSSGSSQP 68
R I IS +KT+ T A++ + GSSQP
Sbjct 1918 RRMIADISDAKTLKTYASQVQMYGGSSQP 1946
> CE10268
Length=412
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 10/36 (27%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 61 LSSGSSQPVKAATTFLGFNSDASCLCVGTTAGFLVW 96
+S+ +S+ + ++GFN D+ +CVG G++ +
Sbjct 1 MSATTSEENPDSINYIGFNQDSKVICVGHKDGYMFY 36
> SPAC458.06
Length=364
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 76 LGFNSDASCLCVGTTAGFLVWSVRPLK 102
+ N DASC+ V G+ ++ + PLK
Sbjct 7 VSLNQDASCMSVALDTGYKIFQINPLK 33
> 7303869
Length=725
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 9/21 (42%), Positives = 14/21 (66%), Gaps = 0/21 (0%)
Query 80 SDASCLCVGTTAGFLVWSVRP 100
S++ C+ +G T G L W+ RP
Sbjct 699 SESHCVAIGATVGSLKWTPRP 719
> CE09739
Length=1394
Score = 27.3 bits (59), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 39 RRSSIGGISSSKTMSTAAAEAGLSSGSSQPVKAATTFLGF 78
R +S+ G TM+T A L++G ++ A T +LGF
Sbjct 1219 RLASVTGFGDEGTMNTDVYNARLTTGDTKTRWARTGYLGF 1258
Lambda K H
0.322 0.131 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40