bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3260_orf1
Length=75
Score E
Sequences producing significant alignments: (Bits) Value
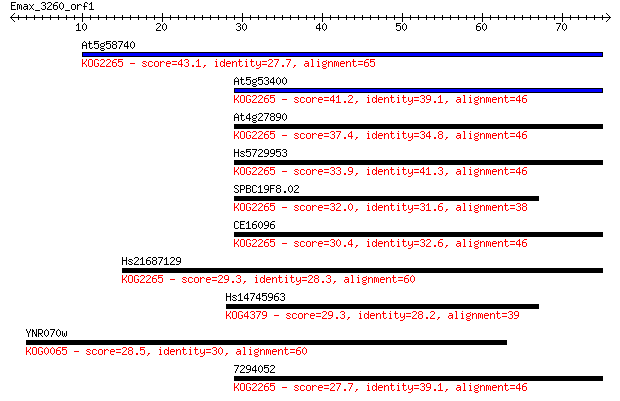
At5g58740 43.1 1e-04
At5g53400 41.2 4e-04
At4g27890 37.4 0.007
Hs5729953 33.9 0.084
SPBC19F8.02 32.0 0.32
CE16096 30.4 0.90
Hs21687129 29.3 1.8
Hs14745963 29.3 1.8
YNR070w 28.5 3.0
7294052 27.7 5.2
> At5g58740
Length=158
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 34/65 (52%), Gaps = 6/65 (9%)
Query 10 DSKYKFIHKLFVMYAGRLLYEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGLIGK 69
+ ++ FIH G+ ++EW+Q L ++++Y++ P K KI + VG+ G
Sbjct 8 EKRHDFIHN------GQKVFEWDQTLEEVNMYITLPPNVHPKSFHCKIQSKHIEVGIKGN 61
Query 70 PPLFD 74
PP +
Sbjct 62 PPYLN 66
> At5g53400
Length=304
Score = 41.2 bits (95), Expect = 4e-04, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 29 YEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGLIGKPPLFD 74
Y W Q+L ++ + + P TKA+ + +I NRL+VGL G+ P+ D
Sbjct 147 YSWIQNLQEVTVNIPVPTGTKARTVVCEIKKNRLKVGLKGQDPIVD 192
> At4g27890
Length=293
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 29 YEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGLIGKPPLFD 74
Y W Q+L ++ + + P TK++ + +I NRL+VGL G+ + D
Sbjct 136 YSWGQNLQEVTINIPMPEGTKSRSVTCEIKKNRLKVGLKGQDLIVD 181
> Hs5729953
Length=331
Score = 33.9 bits (76), Expect = 0.084, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 26/48 (54%), Gaps = 2/48 (4%)
Query 29 YEWEQDLSDIHLYLS--PPAQTKAKDIQVKIHPNRLRVGLIGKPPLFD 74
Y W Q LS++ L + + K KD+ V I LRVGL G+P + D
Sbjct 172 YRWTQTLSELDLAVPFCVNFRLKGKDMVVDIQRRHLRVGLKGQPAIID 219
> SPBC19F8.02
Length=166
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 29 YEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGL 66
YEW+Q ++D+ + + P T+AK +QV + + L++ +
Sbjct 11 YEWDQTIADVDIVIHVPKGTRAKSLQVDMSNHDLKIQI 48
> CE16096
Length=320
Score = 30.4 bits (67), Expect = 0.90, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 26/48 (54%), Gaps = 2/48 (4%)
Query 29 YEWEQDLSDIHLYLSPPA--QTKAKDIQVKIHPNRLRVGLIGKPPLFD 74
Y+W Q L ++ + + A K++D+ VKI + VGL + P+ D
Sbjct 161 YQWTQTLQELEVKIPIAAGFAIKSRDVVVKIEKTSVSVGLKNQAPIVD 208
> Hs21687129
Length=157
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Query 15 FIHKLFVMYAGRLLYEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGLIG----KP 70
F + V+ G +W Q L ++ + + P T+A+DIQ + + + + G K
Sbjct 5 FEERSGVVPCGTPWGQWYQTLEEVFIEVQVPPGTRAQDIQCGLQSRHVALSVGGREILKG 64
Query 71 PLFD 74
LFD
Sbjct 65 KLFD 68
> Hs14745963
Length=583
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 11/39 (28%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 28 LYEWEQDLSDIHLYLSPPAQTKAKDIQVKIHPNRLRVGL 66
LY W+Q D+ + + P + +DIQ++ P+ + + L
Sbjct 277 LYYWQQTEDDLTVTIRLPEDSTKEDIQIQFLPDHINIVL 315
> YNR070w
Length=1333
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 3 MSVQSNVDSKYKFIHKLFVMYAGRLLY----EWEQDLSDIHLYLSPPAQTKAKDIQVKIH 58
++V ++ Y+ K+ V+YAGR ++ +D + YL PP Q+ A+ +
Sbjct 235 VTVYQASENIYETFDKVTVLYAGRQIFCGKTTEAKDYFENMGYLCPPRQSTAEYLTAITD 294
Query 59 PNRL 62
PN L
Sbjct 295 PNGL 298
> 7294052
Length=332
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 18/64 (28%)
Query 29 YEWEQDLSDIHL--------YLSPPAQTK----------AKDIQVKIHPNRLRVGLIGKP 70
Y W Q L ++ + Y S AQ K A+D+ + I L+VG+ G+
Sbjct 173 YTWTQTLEEVEVSPGTSLTPYFSYLAQLKIPFNLTFGLRARDLVISIGKKSLKVGIKGQT 232
Query 71 PLFD 74
P+ D
Sbjct 233 PIID 236
Lambda K H
0.322 0.140 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181410888
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40