bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3374_orf3
Length=68
Score E
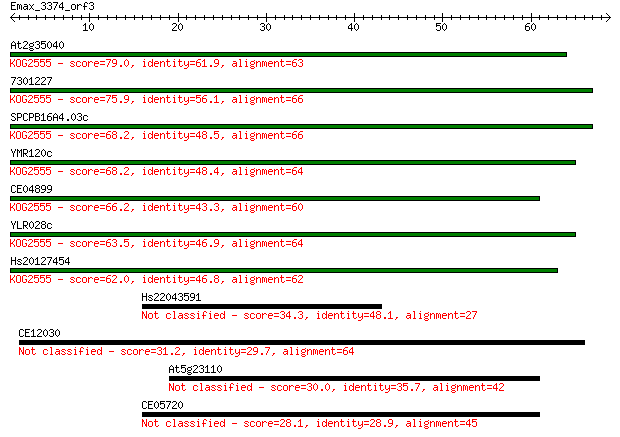
Sequences producing significant alignments: (Bits) Value
At2g35040 79.0 2e-15
7301227 75.9 2e-14
SPCPB16A4.03c 68.2 4e-12
YMR120c 68.2 4e-12
CE04899 66.2 1e-11
YLR028c 63.5 9e-11
Hs20127454 62.0 3e-10
Hs22043591 34.3 0.062
CE12030 31.2 0.54
At5g23110 30.0 1.1
CE05720 28.1 4.0
> At2g35040
Length=545
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 39/64 (60%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 1 VAVNLYPFEATINKPG-CDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLES 59
V VNLYPF + PG IENIDIGGP M+R+AAKNHKDV IVV++ DY VLE
Sbjct 116 VVVNLYPFYEKVTAPGGISFEDGIENIDIGGPAMIRAAAKNHKDVLIVVDSGDYQAVLEY 175
Query 60 LKAG 63
LK G
Sbjct 176 LKGG 179
> 7301227
Length=590
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/66 (56%), Positives = 45/66 (68%), Gaps = 0/66 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF +T+ KP L A+ENIDIGG T++R+AAKNH+ V +V A DY VL L
Sbjct 99 VVCNLYPFASTVAKPDVTLADAVENIDIGGVTLLRAAAKNHQRVTVVCEAVDYDRVLSEL 158
Query 61 KAGGLT 66
KA G T
Sbjct 159 KASGNT 164
> SPCPB16A4.03c
Length=585
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/66 (48%), Positives = 42/66 (63%), Gaps = 0/66 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF TI KP +P A+E IDIGG T++R+AAKNH V I+ + +DYA +
Sbjct 95 VVCNLYPFRETIAKPNVTIPEAVEEIDIGGVTLLRAAAKNHARVTILSDPADYATFTDKF 154
Query 61 KAGGLT 66
+ LT
Sbjct 155 LSDKLT 160
> YMR120c
Length=592
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ K G +P A+E IDIGG T++R+AAKNH V I+ + DY+ L L
Sbjct 100 VVCNLYPFKETVAKVGVTIPEAVEEIDIGGVTLLRAAAKNHARVTILSDPKDYSEFLSEL 159
Query 61 KAGG 64
+ G
Sbjct 160 SSNG 163
> CE04899
Length=267
Score = 66.2 bits (160), Expect = 1e-11, Method: Composition-based stats.
Identities = 26/60 (43%), Positives = 42/60 (70%), Gaps = 0/60 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ C + A+ENIDIGG T++R+AAKNH+ V+++ + +DY +++
Sbjct 121 VVCNLYPFKKTVQSKDCSVEEAVENIDIGGVTLLRAAAKNHERVSVICDPADYDHIISEF 180
> YLR028c
Length=591
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ K G + A+E IDIGG T++R+AAKNH V I+ + +DY+ L+ L
Sbjct 100 VVCNLYPFKETVAKIGVTVQEAVEEIDIGGVTLLRAAAKNHSRVTILSDPNDYSIFLQDL 159
Query 61 KAGG 64
G
Sbjct 160 SKDG 163
> Hs20127454
Length=592
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 39/62 (62%), Gaps = 0/62 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
VA NLYPF T+ PG + A+E IDIGG T++R+AAKNH V +V DY V +
Sbjct 99 VACNLYPFVKTVASPGVTVEEAVEQIDIGGVTLLRAAAKNHARVTVVCEPEDYVVVSTEM 158
Query 61 KA 62
++
Sbjct 159 QS 160
> Hs22043591
Length=405
Score = 34.3 bits (77), Expect = 0.062, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 16 GCDLPTAIENIDIGGPTMVRSAAKNHK 42
GCDLP I +D GPT R A + H+
Sbjct 18 GCDLPIGISALDTQGPTAARHAEQGHR 44
> CE12030
Length=709
Score = 31.2 bits (69), Expect = 0.54, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 10/65 (15%)
Query 2 AVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLE-SL 60
A+N+ F A + K +L NI +GGP +R + + +DYA++ + +L
Sbjct 474 AINMVEFGARLKKLKTELEIWKSNITVGGPHRIRRS---------TTSLADYAHIFQNAL 524
Query 61 KAGGL 65
K GG+
Sbjct 525 KVGGV 529
> At5g23110
Length=4706
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 19 LPTAIENI--DIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
LP A+ N+ IGG + + H D++ V+ + Y VLES+
Sbjct 725 LPAAVRNVLEKIGGKILNNNIKVEHSDLSSFVSDASYTGVLESI 768
> CE05720
Length=429
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 16 GCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
GC P E+ + P+M RS N+ + ++ ++Y V+E L
Sbjct 290 GCVPPIETEHENCQSPSMKRSRCTNYSFRTLTLSTAEYTKVVEFL 334
Lambda K H
0.314 0.132 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1203543208
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40