bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3381_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
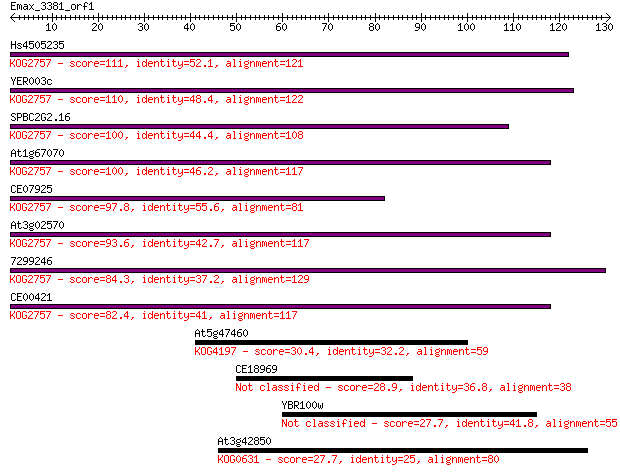
Hs4505235 111 3e-25
YER003c 110 5e-25
SPBC2G2.16 100 5e-22
At1g67070 100 8e-22
CE07925 97.8 5e-21
At3g02570 93.6 8e-20
7299246 84.3 5e-17
CE00421 82.4 2e-16
At5g47460 30.4 0.83
CE18969 28.9 2.4
YBR100w 27.7 5.8
At3g42850 27.7 5.9
> Hs4505235
Length=423
Score = 111 bits (278), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 63/126 (50%), Positives = 79/126 (62%), Gaps = 5/126 (3%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV T LSIQAHPNK LAEKLH P HYPDANHKPE+AIALT F+ CGF+P EI
Sbjct 98 FKVLSVETPLSIQAHPNKELAEKLHLQAPQHYPDANHKPEMAIALTPFQGLCGFRPVEEI 157
Query 61 EFLLNSVPGFKNIIGQDLVA--KYQSAGDKK---ETLKELFSALMTAPEELVKNNLSSLK 115
L VP F+ +IG + K + D + +L+ FS LM + +++V L+ L
Sbjct 158 VTFLKKVPEFQFLIGDEAATHLKQTMSHDSQAVASSLQSCFSHLMKSEKKVVVEQLNLLV 217
Query 116 TRRTTQ 121
R + Q
Sbjct 218 KRISQQ 223
> YER003c
Length=429
Score = 110 bits (276), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 59/134 (44%), Positives = 80/134 (59%), Gaps = 12/134 (8%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLS+ LSIQAHP+KAL + LH DP +YPD NHKPE+AIA+T+FE FCGFKP EI
Sbjct 97 FKVLSIEKVLSIQAHPDKALGKILHAQDPKNYPDDNHKPEMAIAVTDFEGFCGFKPLQEI 156
Query 61 EFLLNSVPGFKNIIGQ----DLVAKYQSAGDK--------KETLKELFSALMTAPEELVK 108
L +P +NI+G+ + + Q + K K+ L+ +FS +M A ++ +K
Sbjct 157 ADELKRIPELRNIVGEETSRNFIENIQPSAQKGSPEDEQNKKLLQAVFSRVMNASDDKIK 216
Query 109 NNLSSLKTRRTTQP 122
SL R P
Sbjct 217 IQARSLVERSKNSP 230
> SPBC2G2.16
Length=412
Score = 100 bits (250), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 75/110 (68%), Gaps = 2/110 (1%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLS+ LSIQAHP+K L ++LH +P Y D NHKPE+A+ALTEF+A CGF+P +I
Sbjct 87 FKVLSINKVLSIQAHPDKPLGKQLHKTNPKEYKDDNHKPEMAVALTEFDALCGFRPVKQI 146
Query 61 EFLLNSVPGFKNIIGQDLVAKYQSA--GDKKETLKELFSALMTAPEELVK 108
E L+S+ + +G++ V +++ D+++ L+ LF+ LM E+ ++
Sbjct 147 EQFLDSIAPLREFVGEEAVRQFKGTVKQDERKALETLFNELMHKDEKRIQ 196
> At1g67070
Length=441
Score = 100 bits (248), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 54/123 (43%), Positives = 75/123 (60%), Gaps = 6/123 (4%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV ALSIQAHPNKALAEKLH +DP Y D NHKPE+A+A+T F+A CGF E+
Sbjct 119 FKVLSVTKALSIQAHPNKALAEKLHREDPLLYRDNNHKPEIALAVTPFQALCGFVTLKEL 178
Query 61 EFLLNSVPGFKNIIGQDL------VAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSL 114
+ ++ +VP ++G V ++ K ++ +F+ LM+A K +S +
Sbjct 179 KEVITNVPEITELVGSKAADQIFNVHEHDEDERIKSVVRLIFTQLMSASNNETKQVVSRM 238
Query 115 KTR 117
K R
Sbjct 239 KNR 241
> CE07925
Length=467
Score = 97.8 bits (242), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 45/81 (55%), Positives = 53/81 (65%), Gaps = 0/81 (0%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
KV+S+RT LS+Q HP K A +LH DP HYPD NHKPELA ALT FE CGF+P EI
Sbjct 89 MKVMSIRTTLSLQVHPTKEQARRLHEKDPIHYPDRNHKPELAYALTRFELLCGFRPAREI 148
Query 61 EFLLNSVPGFKNIIGQDLVAK 81
L + P F+ + G D AK
Sbjct 149 LQNLQTFPSFRLLFGGDTYAK 169
> At3g02570
Length=432
Score = 93.6 bits (231), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 50/122 (40%), Positives = 72/122 (59%), Gaps = 5/122 (4%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV LSIQAHP+K LA+K+H P Y D NHKPE+A+A T+FEA CGF P E+
Sbjct 112 FKVLSVARPLSIQAHPDKKLAKKMHKAHPNLYKDDNHKPEMALAYTQFEALCGFIPLQEL 171
Query 61 EFLLNSVPGFKNIIG-----QDLVAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSLK 115
+ ++ ++P + ++G Q K ++ +F+ LM+A + K +S LK
Sbjct 172 KSVIRAIPEIEELVGSEEANQVFCITEHDEEKVKSVVRTIFTLLMSADADTTKKIVSKLK 231
Query 116 TR 117
R
Sbjct 232 RR 233
> 7299246
Length=396
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 48/129 (37%), Positives = 70/129 (54%), Gaps = 1/129 (0%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLS+ ALSIQ HPNK AE+LH + P Y D NHKPELAIALT F A GF +I
Sbjct 74 FKVLSINKALSIQVHPNKCEAERLHKERPDIYKDPNHKPELAIALTPFLALVGFMSAEDI 133
Query 61 EFLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELFSALMTAPEELVKNNLSSLKTRRTT 120
++ +IG++ V + + + E++K + LM E ++ +S +
Sbjct 134 RDYIDEFQPLSKLIGKEAVDQLHDSTN-SESVKLCYEKLMKTEEPVIAKCISEIAKDYQN 192
Query 121 QPETEPRLE 129
+ ++ LE
Sbjct 193 ELKSNDLLE 201
> CE00421
Length=411
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 48/121 (39%), Positives = 70/121 (57%), Gaps = 9/121 (7%)
Query 1 FKVLSVRTALSIQAHPNKALAEKLHHDDPFHYPDANHKPELAIALTEFEAFCGFKPHAEI 60
FKVLSV LSIQ HP K + LH DP +YPD NHKPE+AIALTEFE GF+ H++I
Sbjct 87 FKVLSVLGPLSIQIHPTKEQGKLLHATDPKNYPDDNHKPEIAIALTEFELLSGFRQHSQI 146
Query 61 EFLLNSVPGFKNIIGQDLVAKYQSAGDKKET----LKELFSALMTAPEELVKNNLSSLKT 116
+ + ++ ++ ++ S G E+ LK++FS + P+E ++ + L
Sbjct 147 MY-----SEIQELLTEEEKSQIDSLGSYGESSAQVLKKIFSRIWRTPKEKLQIVVDKLAR 201
Query 117 R 117
R
Sbjct 202 R 202
> At5g47460
Length=576
Score = 30.4 bits (67), Expect = 0.83, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 3/59 (5%)
Query 41 LAIALTEFEAFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETLKELFSAL 99
+A AL + + CG HAE+ F ++P I+ ++++ Y GD E +K LF+ L
Sbjct 357 VASALIDMYSKCGMLKHAELMFW--TMPRKNLIVWNEMISGYARNGDSIEAIK-LFNQL 412
> CE18969
Length=304
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 50 AFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGD 87
AF GF + + FL+ +P KN G ++ QS GD
Sbjct 9 AFIGFMANWSVAFLIKKLPSLKNSFG--MLTTSQSIGD 44
> YBR100w
Length=112
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 23/66 (34%), Positives = 35/66 (53%), Gaps = 13/66 (19%)
Query 60 IEFLLNSVPGFKNIIGQ---DLVAKY--------QSAGDKKETLKELFSALMTAPEELVK 108
+EF+ NS+P ++IG+ D +Y +SA D ETLK+ F + P E+
Sbjct 1 MEFI-NSLPNLVSLIGKQRMDPAIRYMKYAHLNVKSAQDSTETLKKTFHQIGRMP-EMKA 58
Query 109 NNLSSL 114
NN+ SL
Sbjct 59 NNVVSL 64
> At3g42850
Length=964
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 34/83 (40%), Gaps = 5/83 (6%)
Query 46 TEFEAFCGFKPHAEIEFLLNSVPGFKNIIGQDLVAKYQSAGDKKETL-KELFSALMTAPE 104
T + C PH + +P ++I G++ + KY GD T+ K+ A+M
Sbjct 768 TSLDYLCNLSPHRFQALYASKLP--QSITGEEFLEKYGDHGDSVTTIDKDGTYAIMAPTR 825
Query 105 ELVKNN--LSSLKTRRTTQPETE 125
+ N + + K T P E
Sbjct 826 HPIYENFRVQAFKALLTATPSEE 848
Lambda K H
0.315 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40