bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_3393_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
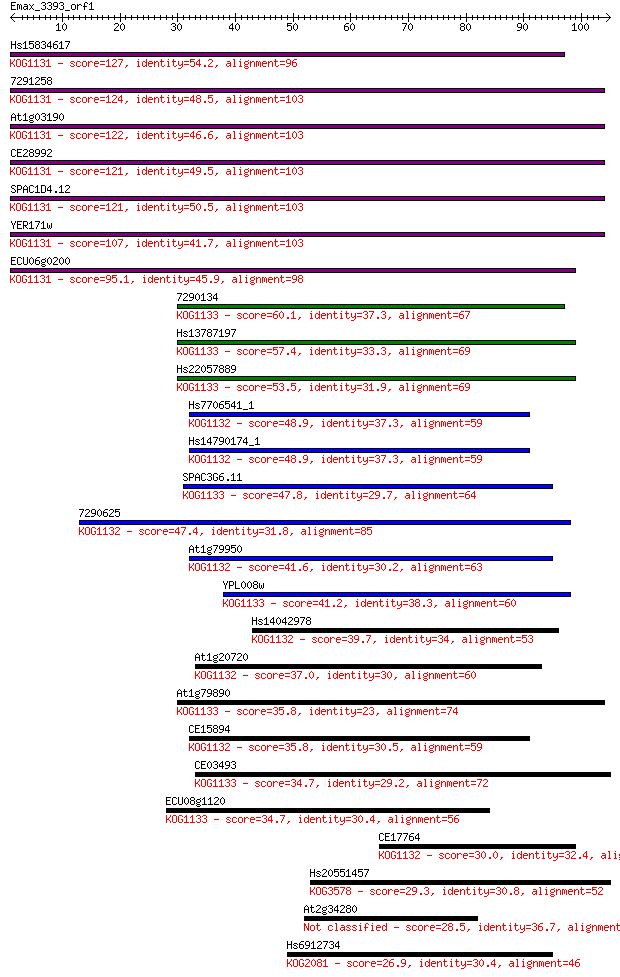
Hs15834617 127 5e-30
7291258 124 4e-29
At1g03190 122 2e-28
CE28992 121 3e-28
SPAC1D4.12 121 3e-28
YER171w 107 4e-24
ECU06g0200 95.1 3e-20
7290134 60.1 9e-10
Hs13787197 57.4 7e-09
Hs22057889 53.5 9e-08
Hs7706541_1 48.9 3e-06
Hs14790174_1 48.9 3e-06
SPAC3G6.11 47.8 5e-06
7290625 47.4 7e-06
At1g79950 41.6 3e-04
YPL008w 41.2 5e-04
Hs14042978 39.7 0.001
At1g20720 37.0 0.009
At1g79890 35.8 0.017
CE15894 35.8 0.018
CE03493 34.7 0.038
ECU08g1120 34.7 0.044
CE17764 30.0 1.2
Hs20551457 29.3 1.9
At2g34280 28.5 3.5
Hs6912734 26.9 8.1
> Hs15834617
Length=760
Score = 127 bits (319), Expect = 5e-30, Method: Composition-based stats.
Identities = 52/96 (54%), Positives = 71/96 (73%), Gaps = 0/96 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YPKIL F P+ +F +++ R C CPMI+ RG DQV ++SKFE R D++V+RNY N
Sbjct 463 PLDIYPKILDFHPVTMATFTMTLARVCLCPMIIGRGNDQVAISSKFETREDIAVIRNYGN 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFL 96
LLL++ VPDG+V FF SY YME+ ++ WYE G L
Sbjct 523 LLLEMSAVVPDGIVAFFTSYQYMESTVASWYEQGIL 558
> 7291258
Length=745
Score = 124 bits (311), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 50/103 (48%), Positives = 76/103 (73%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
P+++YPKIL F P++ SF +++ R C PMIV++G DQV ++SKFE R D +V+RNY
Sbjct 439 PMDMYPKILDFDPVVMSSFTMTLARPCLLPMIVSKGNDQVTISSKFETREDTAVIRNYGQ 498
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL+++ K+VPDG+VCFF SY Y+E+ ++ WY+ G + L +K
Sbjct 499 LLVEVAKTVPDGIVCFFTSYLYLESVVASWYDQGIVDTLLRYK 541
> At1g03190
Length=758
Score = 122 bits (306), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 48/103 (46%), Positives = 76/103 (73%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
P++LYP++L+F P+++ SF +SM R C CPM++ RG+DQ+P+++KF+ R D V+RNY
Sbjct 463 PIDLYPRLLNFTPVVSRSFKMSMTRDCICPMVLTRGSDQLPVSTKFDMRSDPGVVRNYGK 522
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
LL+++ VPDG+VCFF SY+YM+ ++ W E G L + + K
Sbjct 523 LLVEMVSIVPDGVVCFFVSYSYMDGIIATWNETGILKEIMQQK 565
> CE28992
Length=751
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 51/103 (49%), Positives = 72/103 (69%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PLE+YPK+L F P + SF +++ R C P++VARG DQV +TS+FE R DV+V+RNY N
Sbjct 453 PLEMYPKVLDFDPSVIASFTMTLARPCLSPLVVARGNDQVAMTSRFEQRADVAVIRNYGN 512
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
L+L++ VPDG+V FF SY YME + WYE + + + +K
Sbjct 513 LVLEMASLVPDGMVVFFTSYLYMENVIGVWYEQHIIDELMKYK 555
> SPAC1D4.12
Length=772
Score = 121 bits (303), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 52/103 (50%), Positives = 74/103 (71%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YPK+L F ++ ES+ +S+ R CF PM+V RG+DQV ++SKFE R+D SV+RNY N
Sbjct 462 PLDMYPKMLQFNTVMQESYGMSLARNCFLPMVVTRGSDQVAISSKFEARNDPSVVRNYGN 521
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
+L++ K PDGLV FFPSY Y+E+ +S W G L + +K
Sbjct 522 ILVEFSKITPDGLVAFFPSYLYLESIVSSWQSMGILDEVWKYK 564
> YER171w
Length=778
Score = 107 bits (268), Expect = 4e-24, Method: Composition-based stats.
Identities = 43/103 (41%), Positives = 73/103 (70%), Gaps = 0/103 (0%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYAN 60
PL++YP++L+F ++ +S+ +++ + F PMI+ +G+DQV ++S+FE R+D S++RNY +
Sbjct 465 PLDMYPRMLNFKTVLQKSYAMTLAKKSFLPMIITKGSDQVAISSRFEIRNDPSIVRNYGS 524
Query 61 LLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFK 103
+L++ K PDG+V FFPSY YME+ +S W G L + K
Sbjct 525 MLVEFAKITPDGMVVFFPSYLYMESIVSMWQTMGILDEVWKHK 567
> ECU06g0200
Length=742
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 66/116 (56%), Gaps = 18/116 (15%)
Query 1 PLELYPKILSFVPLIAESFPVSMERACFCPMIVARGADQVPL------------------ 42
P+++YPKIL+FVP +++R P+I+ +G DQ+ L
Sbjct 440 PIDMYPKILNFVPSRIAEIGATLDRNSISPLIITKGNDQMTLRALSDDMETTDACSGDVL 499
Query 43 TSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQ 98
T+ F R D SV+RNY +L+++L K VPDG+VCFFPSY YME +S W E + +
Sbjct 500 TTSFSLRSDPSVVRNYGHLMVELSKVVPDGIVCFFPSYMYMEEIVSLWAETSIINE 555
> 7290134
Length=861
Score = 60.1 bits (144), Expect = 9e-10, Method: Composition-based stats.
Identities = 25/67 (37%), Positives = 39/67 (58%), Gaps = 0/67 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P +++ G PL+ KF R +LR + +L +LC+ VP G+VCF PSY Y++ Y
Sbjct 627 PFVISNGPSGAPLSFKFAHRGSAEMLRELSMILRNLCQVVPGGVVCFLPSYEYLDKVYKY 686
Query 90 WYENGFL 96
++G L
Sbjct 687 LEQSGTL 693
> Hs13787197
Length=906
Score = 57.4 bits (137), Expect = 7e-09, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 0/69 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+++ G PL F+ R ++ +L +LC VP G+VCFFPSY Y+ ++
Sbjct 660 PLVICSGISNQPLEFTFQKRELPQMMDEVGRILCNLCGVVPGGVVCFFPSYEYLRQVHAH 719
Query 90 WYENGFLAQ 98
W + G L +
Sbjct 720 WEKGGLLGR 728
> Hs22057889
Length=960
Score = 53.5 bits (127), Expect = 9e-08, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 0/69 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+++ G PL F+ R ++ +L +LC V G+VCFFPSY Y+ ++
Sbjct 660 PLVICSGVSNQPLEFTFQKRDLPQMMDEVGRILCNLCGVVSGGVVCFFPSYEYLRQVHAH 719
Query 90 WYENGFLAQ 98
W + G L +
Sbjct 720 WEKGGLLGR 728
> Hs7706541_1
Length=755
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
+V RG D L+S F+ R L + L ++ + VP GL+ FFPSY ME ++ +W
Sbjct 515 VVPRGPDGAQLSSAFDRRFSEECLSSLGKALGNIARVVPYGLLIFFPSYPVMEKSLEFW 573
> Hs14790174_1
Length=766
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
+V RG D L+S F+ R L + L ++ + VP GL+ FFPSY ME ++ +W
Sbjct 515 VVPRGPDGAQLSSAFDRRFSEECLSSLGKALGNIARVVPYGLLIFFPSYPVMEKSLEFW 573
> SPAC3G6.11
Length=844
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 31 MIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
++V++G VP + + D ++L++ + +PDG+V FFPS+A+++ A+ W
Sbjct 604 ILVSQGPAGVPFEFTHKRKDDENLLKDLGRTFQNFISIIPDGVVVFFPSFAFLQQAVKVW 663
Query 91 YENG 94
NG
Sbjct 664 EMNG 667
> 7290625
Length=985
Score = 47.4 bits (111), Expect = 7e-06, Method: Composition-based stats.
Identities = 27/94 (28%), Positives = 47/94 (50%), Gaps = 9/94 (9%)
Query 13 PLIAE-SFPVS--------MERACFCPMIVARGADQVPLTSKFECRHDVSVLRNYANLLL 63
PLIAE + PV+ ++++ I+ G D+ L S + R + + + +L
Sbjct 488 PLIAELAIPVAQHLENPHIVDQSQVYVKIIGTGPDRQQLISNYANRDNPKYISSLGQTIL 547
Query 64 DLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLA 97
++ + VPDGL+ FFPSY + + W +G A
Sbjct 548 NVARIVPDGLLVFFPSYPMLNKCVDAWQASGLWA 581
> At1g79950
Length=983
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWY 91
+V+ G L S + R + N +++ + VP+GL+ FFPSY M++ +++W
Sbjct 468 VVSTGPSGYVLNSSYRNRDVPEYKQELGNAIVNFSRVVPEGLLIFFPSYYLMDSCITFW- 526
Query 92 ENG 94
+NG
Sbjct 527 KNG 529
> YPL008w
Length=861
Score = 41.2 bits (95), Expect = 5e-04, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Query 38 DQVPLTSKFECRHDVSVLRNYA-NLLLDLCKSVPD--GLVCFFPSYAYMETAMSYWYENG 94
+Q L FE R S++ N+ +DL K+VP G+V FFPSY Y+ + W +N
Sbjct 625 NQPELEFTFEKRMSPSLVNNHLFQFFVDLSKAVPKKGGIVAFFPSYQYLAHVIQCWKQND 684
Query 95 FLA 97
A
Sbjct 685 RFA 687
> Hs14042978
Length=1249
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 6/53 (11%)
Query 43 TSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGF 95
T FE + +V L LL +C++V G++CF PSY +E W G
Sbjct 667 TETFEFQDEVGAL------LLSVCQTVSQGILCFLPSYKLLEKLKERWLSTGL 713
> At1g20720
Length=986
Score = 37.0 bits (84), Expect = 0.009, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 26/60 (43%), Gaps = 0/60 (0%)
Query 33 VARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYE 92
++ G PL + ++ S L ++C VP G + FFPSY ME W E
Sbjct 491 ISNGPSNYPLNASYKTADAYSFQDALGKSLEEICTIVPGGSLVFFPSYKLMEKLCMRWRE 550
> At1g79890
Length=882
Score = 35.8 bits (81), Expect = 0.017, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 33/74 (44%), Gaps = 0/74 (0%)
Query 30 PMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSY 89
P+ V+ G R + +++ L+ +L VP+G++ FF S+ Y +
Sbjct 602 PIAVSHGPSGQSFDFSHSSRSSIGMIQELGLLMSNLVAVVPEGIIVFFSSFEYETQVHTA 661
Query 90 WYENGFLAQTLPFK 103
W +G L + + K
Sbjct 662 WSNSGILRRIVKKK 675
> CE15894
Length=994
Score = 35.8 bits (81), Expect = 0.018, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 32 IVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYW 90
IV RG + L F+ R ++ + A LL + + +P G++ FF SY+ M+ ++ W
Sbjct 527 IVTRG-KRGGLAGSFQNRKNLDYVTGVAEALLRVMEVIPQGILIFFSSYSQMDELVATW 584
> CE03493
Length=848
Score = 34.7 bits (78), Expect = 0.038, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 0/72 (0%)
Query 33 VARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYE 92
V R D P ++ R + LR+ A + L +P+G+V F PSY ++ E
Sbjct 621 VERTVDGKPFQLTYQTRGADTTLRSLATSIQALIPHIPNGVVIFVPSYDFLFNFQKKMKE 680
Query 93 NGFLAQTLPFKA 104
G L + KA
Sbjct 681 FGILKRIEEKKA 692
> ECU08g1120
Length=619
Score = 34.7 bits (78), Expect = 0.044, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 28 FCPMIVARGADQVPLTSKFECRHDVSVLRNYANLLLDLCKSVPDG-LVCFFPSYAYM 83
F ++V G + FE R +++ + + +L +V DG +VCF PS AY+
Sbjct 408 FLALVVGSGPSGREIIVNFETRERPESIKDVTSSISNLSNAVKDGGMVCFLPSKAYL 464
> CE17764
Length=983
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 65 LCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQ 98
+C +VP G++CF PSY ++ N + Q
Sbjct 758 VCSNVPAGILCFLPSYRVLDQLKQCMIRNSTMRQ 791
> Hs20551457
Length=1173
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 24/52 (46%), Gaps = 10/52 (19%)
Query 53 SVLRNYANLLLDLCKSVPDGLVCFFPSYAYMETAMSYWYENGFLAQTLPFKA 104
++ + + LLD+CK VP A TA W+ + FL Q +P A
Sbjct 331 TIDKKFYKSLLDICKKVP----------AITLTANIIWFPDNFLIQKIPAAA 372
> At2g34280
Length=391
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 52 VSVLRNYANLLLDLCKSVPDGLVCFFPSYA 81
V +L + + L +C+S DGL+C + +YA
Sbjct 86 VRILTPWEDTLYKVCQSSCDGLICLYNTYA 115
> Hs6912734
Length=923
Score = 26.9 bits (58), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 14/65 (21%), Positives = 26/65 (40%), Gaps = 19/65 (29%)
Query 49 RHDVSVLRNYANLLLDLCKSVPDGLVCF-------------------FPSYAYMETAMSY 89
R D+ + NY + +LC++ + +VC P Y +E + ++
Sbjct 286 REDLDKVLNYCRIFTELCETFLEKIVCTPGQGLGDLRTLELLLICAGHPQYEVVEISFNF 345
Query 90 WYENG 94
WY G
Sbjct 346 WYRLG 350
Lambda K H
0.327 0.140 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40