bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0012_orf1
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
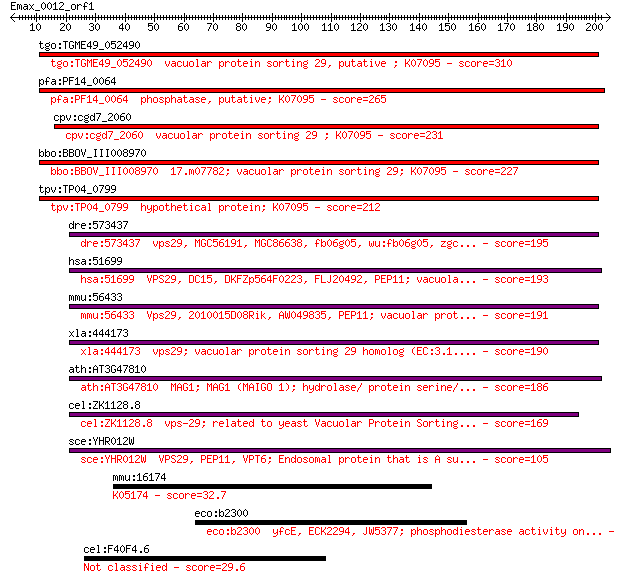
tgo:TGME49_052490 vacuolar protein sorting 29, putative ; K07095 310 2e-84
pfa:PF14_0064 phosphatase, putative; K07095 265 6e-71
cpv:cgd7_2060 vacuolar protein sorting 29 ; K07095 231 1e-60
bbo:BBOV_III008970 17.m07782; vacuolar protein sorting 29; K07095 227 2e-59
tpv:TP04_0799 hypothetical protein; K07095 212 6e-55
dre:573437 vps29, MGC56191, MGC86638, fb06g05, wu:fb06g05, zgc... 195 1e-49
hsa:51699 VPS29, DC15, DKFZp564F0223, FLJ20492, PEP11; vacuola... 193 3e-49
mmu:56433 Vps29, 2010015D08Rik, AW049835, PEP11; vacuolar prot... 191 2e-48
xla:444173 vps29; vacuolar protein sorting 29 homolog (EC:3.1.... 190 4e-48
ath:AT3G47810 MAG1; MAG1 (MAIGO 1); hydrolase/ protein serine/... 186 5e-47
cel:ZK1128.8 vps-29; related to yeast Vacuolar Protein Sorting... 169 6e-42
sce:YHR012W VPS29, PEP11, VPT6; Endosomal protein that is A su... 105 1e-22
mmu:16174 Il18rap; interleukin 18 receptor accessory protein; ... 32.7 1.00
eco:b2300 yfcE, ECK2294, JW5377; phosphodiesterase activity on... 30.8 3.4
cel:F40F4.6 hypothetical protein 29.6 7.5
> tgo:TGME49_052490 vacuolar protein sorting 29, putative ; K07095
Length=203
Score = 310 bits (795), Expect = 2e-84, Method: Compositional matrix adjust.
Identities = 138/190 (72%), Positives = 165/190 (86%), Gaps = 0/190 (0%)
Query 11 MAGNFTDFGDLVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRG 70
M NFT+FGDLVL+IGDFH+PQRAVDLPPCF+ELL+TDKIRHVLCTGNVG SV++ LR
Sbjct 1 MGANFTEFGDLVLLIGDFHIPQRAVDLPPCFRELLNTDKIRHVLCTGNVGCASVVDSLRS 60
Query 71 IAGSVHIVKGDMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIV 130
I+ S+HIVKGD D G DFP YK+LQFG+FKV L+HGHQ++P+GD +LL WQR+LDCDI+
Sbjct 61 ISSSLHIVKGDADAGFDFPEYKVLQFGQFKVGLIHGHQIVPYGDGGSLLHWQRKLDCDIL 120
Query 131 VSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGK 190
V GH H +SV E+EG+FF+NPGSATGAYQPW PSFMLMA+QG+SVVLY+YEE++GK
Sbjct 121 VYGHLHKDSVVELEGKFFVNPGSATGAYQPWLTEKVPSFMLMAVQGSSVVLYVYEEKNGK 180
Query 191 AEVVMSEFSK 200
AEVVMSEF K
Sbjct 181 AEVVMSEFKK 190
> pfa:PF14_0064 phosphatase, putative; K07095
Length=194
Score = 265 bits (678), Expect = 6e-71, Method: Compositional matrix adjust.
Identities = 116/192 (60%), Positives = 147/192 (76%), Gaps = 0/192 (0%)
Query 11 MAGNFTDFGDLVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRG 70
M+G D G+LVL+IGDFH P R + LP CFKELL TDKI+HVLCTGNVG LE L+
Sbjct 1 MSGKLEDIGELVLLIGDFHSPIRNLGLPDCFKELLKTDKIKHVLCTGNVGCNENLELLKN 60
Query 71 IAGSVHIVKGDMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIV 130
IA SVHI KGDMD+ DFP L G+FK++L+HGHQ+IPWGD +ALLQWQ++ D DI+
Sbjct 61 IADSVHITKGDMDDNFDFPEDITLCIGDFKISLIHGHQIIPWGDMNALLQWQKKYDSDII 120
Query 131 VSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGK 190
+SGHTH NS+ + EG++FINPGS TGA+QPW PTP+F+LMA+ +++VLY+YEE++GK
Sbjct 121 ISGHTHKNSIVQYEGKYFINPGSVTGAFQPWLSEPTPTFILMAVAKSNIVLYVYEEKNGK 180
Query 191 AEVVMSEFSKPA 202
V MSE K
Sbjct 181 TNVEMSELHKST 192
> cpv:cgd7_2060 vacuolar protein sorting 29 ; K07095
Length=197
Score = 231 bits (588), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 110/194 (56%), Positives = 137/194 (70%), Gaps = 9/194 (4%)
Query 16 TDFGDLVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSV 75
TDFGDLVL+IGD +P A +LP F+ELL TDKI +VLCTGNV S+ +E L+ I +V
Sbjct 4 TDFGDLVLLIGDLKIPYGAKELPSNFRELLATDKINYVLCTGNVCSQEYVEMLKNITKNV 63
Query 76 HIVKGDMDEGM---------DFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLD 126
+IV GD+D + FP Y ++Q GEFK+ L+HG+QV+PW D +L QWQRRLD
Sbjct 64 YIVSGDLDSAIFNPDPESNGVFPEYVVVQIGEFKIGLMHGNQVLPWDDPGSLEQWQRRLD 123
Query 127 CDIVVSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEE 186
CDI+V+GHTH V E G+ F+NPG+ATGA+ P PSFMLMALQG VVLY+Y+
Sbjct 124 CDILVTGHTHKLRVFEKNGKLFLNPGTATGAFSALTPDAPPSFMLMALQGNKVVLYVYDL 183
Query 187 RDGKAEVVMSEFSK 200
RDGK V MSEFSK
Sbjct 184 RDGKTNVAMSEFSK 197
> bbo:BBOV_III008970 17.m07782; vacuolar protein sorting 29; K07095
Length=215
Score = 227 bits (578), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 95/190 (50%), Positives = 143/190 (75%), Gaps = 0/190 (0%)
Query 11 MAGNFTDFGDLVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRG 70
M+ D G+L++++GD HVPQRA+DLP CF++LL+TDKI+ VLCTGNVGS+ + + L G
Sbjct 1 MSNGGGDLGELLMLVGDLHVPQRALDLPQCFRDLLNTDKIKQVLCTGNVGSQQMKDLLLG 60
Query 71 IAGSVHIVKGDMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIV 130
I+ ++H+VKGD D+ P I+ G FK+ L++G+Q+ WGD +A+ ++ + D D++
Sbjct 61 ISPNLHMVKGDFDQDTTLPEELIIHVGNFKIGLINGYQLPSWGDKNAVYEYAKNRDVDVL 120
Query 131 VSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGK 190
V GHTH + V ++ G+ +NPGSATGA+QPWAP+ P+FMLMA+QG+ +V+Y+YEE +G+
Sbjct 121 VYGHTHISDVSKISGKILVNPGSATGAFQPWAPNAIPTFMLMAVQGSKIVIYVYEEHEGQ 180
Query 191 AEVVMSEFSK 200
A VVMSE +
Sbjct 181 ANVVMSEVDQ 190
> tpv:TP04_0799 hypothetical protein; K07095
Length=213
Score = 212 bits (540), Expect = 6e-55, Method: Compositional matrix adjust.
Identities = 91/190 (47%), Positives = 135/190 (71%), Gaps = 0/190 (0%)
Query 11 MAGNFTDFGDLVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRG 70
M + D G+L++++GD HVPQR++ LPPCFK LL TDKI+ V+CTGNVGS +LE L
Sbjct 1 MGNDEHDLGELLMVLGDLHVPQRSLFLPPCFKRLLKTDKIKRVICTGNVGSNEMLEVLND 60
Query 71 IAGSVHIVKGDMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIV 130
I+ S+HIV+GD D+ + P + G+ K+ +++G+Q+ W + D LL+ ++ DI+
Sbjct 61 ISPSLHIVQGDYDDDFNHPETLTINVGDLKIGVINGYQIPTWNNKDLLLKVAVDMNVDIL 120
Query 131 VSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGK 190
V GH+H + + + G+ F+NPGSATG YQPW P P+FMLMA+QG+ VV+Y+YEE DG+
Sbjct 121 VYGHSHVSDISKHGGKIFVNPGSATGCYQPWQPKSIPTFMLMAIQGSKVVIYVYEEHDGE 180
Query 191 AEVVMSEFSK 200
A+V+M+E
Sbjct 181 AQVIMTELDN 190
> dre:573437 vps29, MGC56191, MGC86638, fb06g05, wu:fb06g05, zgc:56191,
zgc:86638; vacuolar protein sorting 29 (yeast) (EC:3.1.3.3);
K07095
Length=182
Score = 195 bits (495), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 88/180 (48%), Positives = 125/180 (69%), Gaps = 0/180 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 2 LVLVLGDLHIPHRCNTLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+LD DI++SGHTH
Sbjct 62 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQLDVDILISGHTHKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + TPSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 122 FENENKFYINPGSATGAYSALESNITPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 181
> hsa:51699 VPS29, DC15, DKFZp564F0223, FLJ20492, PEP11; vacuolar
protein sorting 29 homolog (S. cerevisiae) (EC:3.1.3.3);
K07095
Length=182
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 87/181 (48%), Positives = 124/181 (68%), Gaps = 0/181 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 2 LVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+ D DI++SGHTH
Sbjct 62 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQFDVDILISGHTHKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + PSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 122 FEHENKFYINPGSATGAYNALETNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 181
Query 201 P 201
P
Sbjct 182 P 182
> mmu:56433 Vps29, 2010015D08Rik, AW049835, PEP11; vacuolar protein
sorting 29 (S. pombe) (EC:3.1.3.3); K07095
Length=182
Score = 191 bits (485), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 86/180 (47%), Positives = 123/180 (68%), Gaps = 0/180 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 2 LVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESYDYLKTLAGDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+ D DI++SGHTH
Sbjct 62 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQFDVDILISGHTHKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + PSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 122 FEHENKFYINPGSATGAYNALETNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 181
> xla:444173 vps29; vacuolar protein sorting 29 homolog (EC:3.1.3.3);
K07095
Length=182
Score = 190 bits (482), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 86/180 (47%), Positives = 123/180 (68%), Gaps = 0/180 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL++GD H+P R LP FK+LL KI+H+LCTGN+ ++ ++L+ +AG VHIV+G
Sbjct 2 LVLVLGDLHIPHRCNSLPAKFKKLLVPGKIQHILCTGNLCTKESFDYLKTLAGDVHIVRG 61
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
D DE +++P K++ G+FK+ L+HGHQVIPWGD +L QR+LD DI++SGHT
Sbjct 62 DFDENLNYPEQKVVTVGQFKIGLIHGHQVIPWGDMASLALLQRQLDVDILISGHTQKFEA 121
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
E E +F+INPGSATGAY + PSF+LM +Q ++VV Y+Y+ +V E+ K
Sbjct 122 FEHENKFYINPGSATGAYNALENNIIPSFVLMDIQASTVVTYVYQLIGDDVKVERIEYKK 181
> ath:AT3G47810 MAG1; MAG1 (MAIGO 1); hydrolase/ protein serine/threonine
phosphatase; K07095
Length=190
Score = 186 bits (472), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 90/181 (49%), Positives = 119/181 (65%), Gaps = 0/181 (0%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL +GD HVP RA DLPP FK +L KI+H++CTGN+ + + ++L+ I +HIV+G
Sbjct 3 LVLALGDLHVPHRAADLPPKFKSMLVPGKIQHIICTGNLCIKEIHDYLKTICPDLHIVRG 62
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSV 140
+ DE +P K L G+FK+ L HGHQVIPWGD D+L QR+L DI+V+GHTH +
Sbjct 63 EFDEDARYPENKTLTIGQFKLGLCHGHQVIPWGDLDSLAMLQRQLGVDILVTGHTHQFTA 122
Query 141 REVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVLYIYEERDGKAEVVMSEFSK 200
+ EG INPGSATGAY PSF+LM + G V+Y+YE DG+ +V EF K
Sbjct 123 YKHEGGVVINPGSATGAYSSINQDVNPSFVLMDIDGFRAVVYVYELIDGEVKVDKIEFKK 182
Query 201 P 201
P
Sbjct 183 P 183
> cel:ZK1128.8 vps-29; related to yeast Vacuolar Protein Sorting
factor family member (vps-29); K07095
Length=187
Score = 169 bits (428), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 79/175 (45%), Positives = 119/175 (68%), Gaps = 2/175 (1%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKG 80
LVL+IGDF++P RA ++ P F++LL +K++HVLCTGN+ S ++LR ++ VHIV+G
Sbjct 2 LVLLIGDFNLPHRAANISPKFRKLLVPNKMQHVLCTGNLCSRETFDYLRTLSSDVHIVRG 61
Query 81 DMD-EGMDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNS 139
+ D E + +P K++ G+F++ + HGHQ+IPWGD L R+LD DI+V+G+T+ S
Sbjct 62 EFDDETLKYPDTKVVTVGQFRIGVCHGHQIIPWGDQRMLELLARQLDVDILVTGNTYECS 121
Query 140 VREVEGRFFINPGSATGAYQPWAPSP-TPSFMLMALQGASVVLYIYEERDGKAEV 193
E GRFF++PGSATG++ P TPSF L+ +Q +VV Y+Y D +V
Sbjct 122 AVEKNGRFFVDPGSATGSFSVTKTEPTTPSFALLDVQADNVVTYLYRLIDDAVKV 176
> sce:YHR012W VPS29, PEP11, VPT6; Endosomal protein that is A
subunit of the membrane-associated retromer complex essential
for endosome-to-Golgi retrograde transport; forms a subcomplex
with Vps35p and Vps26p that selects cargo proteins for
endosome-to-Golgi retrieval; K07095
Length=282
Score = 105 bits (262), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 68/203 (33%), Positives = 100/203 (49%), Gaps = 38/203 (18%)
Query 21 LVLIIGDFHVPQRAVDLPPCFKELLHT-DKIRHVLCTGN-VGSESVLEFLRGIAGSVHIV 78
L+L + D H+P RA DLP FK+LL DKI V GN S L+F+ I+ ++ IV
Sbjct 2 LLLALSDAHIPDRATDLPVKFKKLLSVPDKISQVALLGNSTKSYDFLKFVNQISNNITIV 61
Query 79 KGDMDEG-----------------MDFPVYKILQFGEFKVALLHGHQVIPWGDADALLQW 121
+G+ D G + P+ I++ G K+ G+ V+P D +LL
Sbjct 62 RGEFDNGHLPSTKKDKASDNSRPMEEIPMNSIIRQGALKIGCCSGYTVVPKNDPLSLLAL 121
Query 122 QRRLDCDIVVSGHTHSNSVREVEGRFFINPGSATGAYQPWAPSPTPSFMLMALQGASVVL 181
R+LD DI++ G TH+ +EG+FF+NPGS TGA+ P ++
Sbjct 122 ARQLDVDILLWGGTHNVEAYTLEGKFFVNPGSCTGAFNTDWP----------------IV 165
Query 182 YIYEERDGKAEVVMSEFSKPAKK 204
+ E+ D E V SE KP K+
Sbjct 166 FDVEDSD---EAVTSEVDKPTKE 185
> mmu:16174 Il18rap; interleukin 18 receptor accessory protein;
K05174 interleukin 18 receptor accessory protein
Length=614
Score = 32.7 bits (73), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 41/108 (37%), Gaps = 6/108 (5%)
Query 36 DLPPCFKELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHIVKGDMDEGMDFPVYKILQ 95
D+ C K +L R+V C E VL L G GS+H + P +
Sbjct 135 DMACCIKTVLEVKPQRNVSCGNTAQDEQVL--LLGSTGSIHCPSLSCQSDVQSPEMTWYK 192
Query 96 FGEFKVALLHGHQVIPWGDADALLQWQRRLDCDIVVSGHTHSNSVREV 143
G LL H+ P AD + Q CD S + S +VR V
Sbjct 193 DGR----LLPEHKKNPIEMADIYVFNQGLYVCDYTQSDNVSSWTVRAV 236
> eco:b2300 yfcE, ECK2294, JW5377; phosphodiesterase activity
on bis-pNPP; K07095
Length=184
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 46/100 (46%), Gaps = 15/100 (15%)
Query 64 VLEFLRGIAGSVHIVKGDMDEGMD-----FPV---YKILQFGEFKVALLHGHQVIPWGDA 115
V E L +A V V+G+ D +D FP+ ++ + + ++ L HGH P +
Sbjct 56 VAERLNEVAHKVIAVRGNCDSEVDQMLLHFPITAPWQQVLLEKQRLFLTHGHLFGP-ENL 114
Query 116 DALLQWQRRLDCDIVVSGHTHSNSVREVEGRFFINPGSAT 155
AL Q D++V GHTH + F NPGS +
Sbjct 115 PALNQ------NDVLVYGHTHLPVAEQRGEIFHFNPGSVS 148
> cel:F40F4.6 hypothetical protein
Length=2214
Score = 29.6 bits (65), Expect = 7.5, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 32/87 (36%), Gaps = 5/87 (5%)
Query 26 GDFHVPQRAVDLPPCFK-ELLHTDKIRHVLCTGNVGSESVLEFLRGIAGSVHI----VKG 80
GD P ++ LP F LH +++L SE + S+H+ G
Sbjct 159 GDLIFPDESIQLPTMFTANYLHLPPGQYMLGPRADTSEQFCTMMMSSRSSIHVSGGFTSG 218
Query 81 DMDEGMDFPVYKILQFGEFKVALLHGH 107
D E D+P K F V +H
Sbjct 219 DQAERSDYPNLKFTYFDTESVVAIHAQ 245
Lambda K H
0.323 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6253620652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40