bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0074_orf2
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
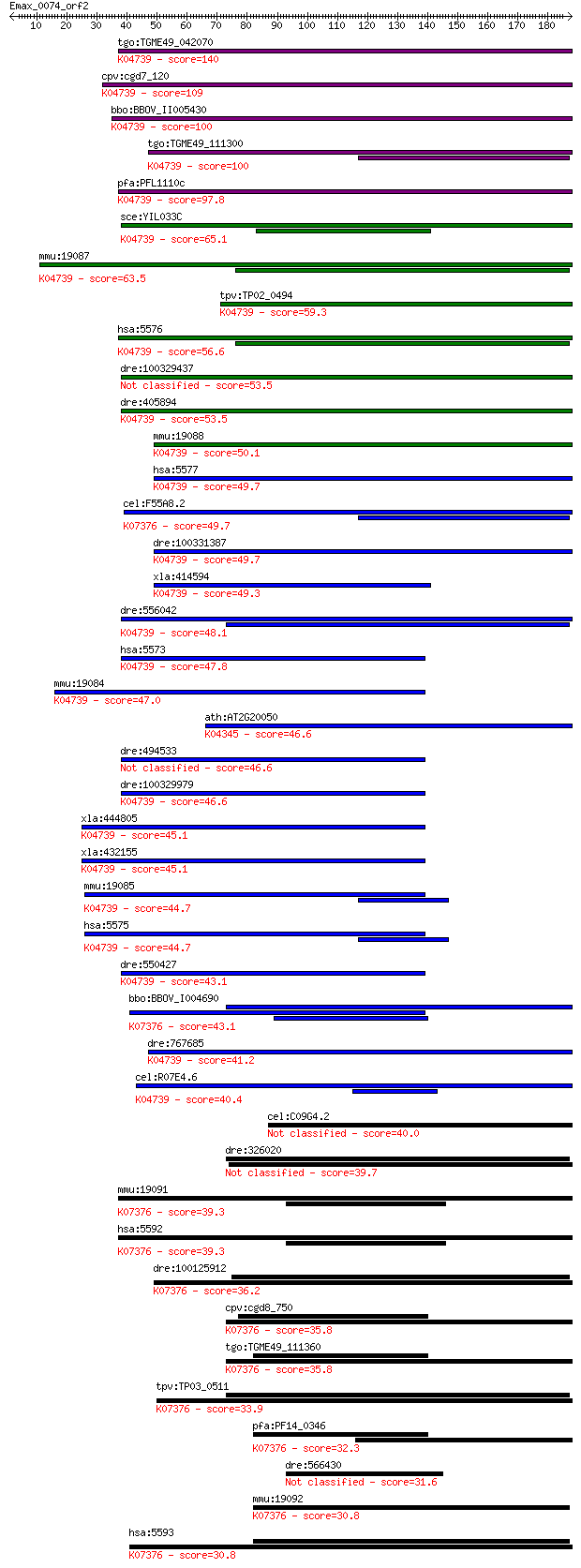
tgo:TGME49_042070 cAMP-dependent protein kinase regulatory sub... 140 2e-33
cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit ... 109 5e-24
bbo:BBOV_II005430 18.m06452; cAMP-dependent protein kinase reg... 100 2e-21
tgo:TGME49_111300 cAMP-dependent protein kinase regulatory sub... 100 3e-21
pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory su... 97.8 2e-20
sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein k... 65.1 1e-10
mmu:19087 Prkar2a, 1110061A24Rik, AI317181, AI836829, RII(alph... 63.5 4e-10
tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit... 59.3 7e-09
hsa:5576 PRKAR2A, MGC3606, PKR2, PRKAR2; protein kinase, cAMP-... 56.6 5e-08
dre:100329437 protein kinase, cAMP-dependent, regulatory, type... 53.5 5e-07
dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kin... 53.5 5e-07
mmu:19088 Prkar2b, AI451071, AW061005, Pkarb2, RII(beta); prot... 50.1 4e-06
hsa:5577 PRKAR2B, PRKAR2, RII-BETA; protein kinase, cAMP-depen... 49.7 5e-06
cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);... 49.7 6e-06
dre:100331387 cAMP-dependent protein kinase, regulatory subuni... 49.7 6e-06
xla:414594 prkar2b, MGC83177; protein kinase, cAMP-dependent, ... 49.3 8e-06
dre:556042 prkar2aa, MGC153742, wu:fb55f12, wu:fj29e12, wu:fj3... 48.1 2e-05
hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR... 47.8 2e-05
mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protei... 47.0 4e-05
ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regu... 46.6 4e-05
dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein k... 46.6 5e-05
dre:100329979 protein kinase, cAMP-dependent, regulatory, type... 46.6 5e-05
xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dep... 45.1 1e-04
xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regu... 45.1 2e-04
mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP depe... 44.7 2e-04
hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regu... 44.7 2e-04
dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependen... 43.1 5e-04
bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:... 43.1 5e-04
dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAM... 41.2 0.002
cel:R07E4.6 kin-2; protein KINase family member (kin-2); K0473... 40.4 0.004
cel:C09G4.2 hypothetical protein 40.0 0.004
dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5 39.7 0.005
mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b; p... 39.3 0.008
hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK, P... 39.3 0.008
dre:100125912 prkg2; protein kinase, cGMP-dependent, type II (... 36.2 0.071
cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase... 35.8 0.082
tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:... 35.8 0.083
tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07... 33.9 0.30
pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12... 32.3 0.96
dre:566430 cGMP dependent protein kinase I 31.6 1.7
mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II; p... 30.8 2.5
hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,... 30.8 2.6
> tgo:TGME49_042070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.10.2); K04739 cAMP-dependent protein
kinase regulator
Length=308
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 75/153 (49%), Positives = 96/153 (62%), Gaps = 23/153 (15%)
Query 37 LPMPSAKV--QAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSL 94
+P PS +AKNRQSVSAE YGEWNK+ FV P YPKT E+K+RI ++E+SFL SSL
Sbjct 1 MPQPSEAYMSRAKNRQSVSAEAYGEWNKRKKFVAPVYPKTAEQKERITKVIESSFLFSSL 60
Query 95 EAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSV 154
+ EDLE ++ AF+E+ V K T II QG DGD+LYLIE GE +V KK P
Sbjct 61 DIEDLETVINAFQEVSVKKGTVIIRQGDDGDRLYLIETGEVDVMKKF-PG---------- 109
Query 155 QQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+++ + + M G+ FGELALMY
Sbjct 110 ----------EKENKFLCKMHPGDAFGELALMY 132
> cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit
; K04739 cAMP-dependent protein kinase regulator
Length=345
Score = 109 bits (273), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 61/156 (39%), Positives = 90/156 (57%), Gaps = 22/156 (14%)
Query 32 DVVDDLPMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLI 91
DV D++ +P + R SVSAE YG WNK F PP YPKT+E+++RIR+ + SF+
Sbjct 41 DVRDNIEIPKNFLARGPRTSVSAEAYGAWNKMKDFTPPSYPKTKEQEKRIREKLLESFMF 100
Query 92 SSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDK 151
+SL+ ++L+ ++ A E V KDT II QG +GDKLY+I++G YKK S
Sbjct 101 TSLDDDELKTVILACVETSVKKDTEIITQGDNGDKLYIIDQGVVECYKKTST-------- 152
Query 152 KSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ ++ L + + G+ FGELAL+Y
Sbjct 153 -------------EPRKHLCD-LNPGDAFGELALLY 174
> bbo:BBOV_II005430 18.m06452; cAMP-dependent protein kinase regulatory
subunit; K04739 cAMP-dependent protein kinase regulator
Length=317
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/157 (38%), Positives = 85/157 (54%), Gaps = 25/157 (15%)
Query 35 DDLPMPSAKV----QAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFL 90
DD+ SA+ +A+NR S+SAEV+G +N FV P + KT E+ RI++ V FL
Sbjct 4 DDIDAISAEAIRFARARNRVSISAEVFGAYNNPDLFVAPVHEKTPEQATRIKETVLKCFL 63
Query 91 ISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQD 150
SS++A+D++ L+ AF+ V T +I+QG GDKLYLIE G A + K S + +Q D
Sbjct 64 FSSVDAKDIDTLVKAFESSDVSAGTKVIQQGDPGDKLYLIESGTAR-FTKTSAATEQVHD 122
Query 151 KKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ + G FGELALMY
Sbjct 123 LGTAGE--------------------GGCFGELALMY 139
> tgo:TGME49_111300 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.11.12); K04739 cAMP-dependent protein
kinase regulator
Length=410
Score = 100 bits (249), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 55/141 (39%), Positives = 81/141 (57%), Gaps = 23/141 (16%)
Query 47 KNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAF 106
K R SVSAEVYGEWNKK +FV P Y K E +K+++ I+ SFL +SL+ +DL ++ A
Sbjct 122 KMRCSVSAEVYGEWNKKKNFVAPVYEKDEGQKEQLERILRQSFLFNSLDEKDLNTVILAM 181
Query 107 KELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQ 166
+E K+ T +I +G DG+ LY+++ GE N K I
Sbjct 182 QEKKIEASTCLIREGDDGECLYIVQSGELNCSKLI-----------------------DG 218
Query 167 QQQLVNVMVAGELFGELALMY 187
++++V V+ G+ FGELAL+Y
Sbjct 219 EERVVKVVGPGDAFGELALLY 239
Score = 33.9 bits (76), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 29/70 (41%), Gaps = 21/70 (30%)
Query 117 IIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVA 176
I+ QG GD Y++EEG A K P Q + KK A
Sbjct 314 IVRQGELGDVFYIVEEGSAVATKSFGPGQPPIEVKK---------------------YQA 352
Query 177 GELFGELALM 186
G+ FGELAL+
Sbjct 353 GDYFGELALI 362
> pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory
subunit, putative; K04739 cAMP-dependent protein kinase regulator
Length=441
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 56/152 (36%), Positives = 85/152 (55%), Gaps = 23/152 (15%)
Query 37 LPMPSAKVQAKNRQSVSAEVYGEWNKKT-HFVPPFYPKTEEEKQRIRDIVENSFLISSLE 95
L + S K R SVSAE YG+WNKK +F+P Y K E+EK +IR+ + SFL + L
Sbjct 135 LDLESIHFIQKKRLSVSAEAYGDWNKKIDNFIPKVYKKDEKEKAKIREALNESFLFNHLN 194
Query 96 AEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQ 155
++ E+++ AF + V K II +G GD LY+I++GE +YK ++ +KK V
Sbjct 195 KKEFEIIVNAFFDKNVEKGVNIINEGDYGDLLYVIDQGEVEIYK------TKENNKKEV- 247
Query 156 QNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ V+ + ++FGELAL+Y
Sbjct 248 ---------------LTVLKSKDVFGELALLY 264
> sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein
kinase regulator
Length=416
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 47/151 (31%), Positives = 71/151 (47%), Gaps = 30/151 (19%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYP-KTEEEKQRIRDIVENSFLISSLEA 96
P+P A+ R SVS E + N + P Y K+E++ QR+ + N+FL + L++
Sbjct 133 PLP-MHFNAQRRTSVSGETL-QPNNFDDWTPDHYKEKSEQQLQRLEKSIRNNFLFNKLDS 190
Query 97 EDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQ 156
+ +++ +E VPK TII+QG GD Y++E+G + Y V
Sbjct 191 DSKRLVINCLEEKSVPKGATIIKQGDQGDYFYVVEKGTVDFY---------------VND 235
Query 157 NEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
N+ VN G FGELALMY
Sbjct 236 NK------------VNSSGPGSSFGELALMY 254
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 83 DIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKK 140
D++++ ++ SL D L A TII +G G+ YLIE G +V KK
Sbjct 295 DLLKSMPVLKSLTTYDRAKLADALDTKIYQPGETIIREGDQGENFYLIEYGAVDVSKK 352
> mmu:19087 Prkar2a, 1110061A24Rik, AI317181, AI836829, RII(alpha);
protein kinase, cAMP dependent regulatory, type II alpha
(EC:2.7.11.1); K04739 cAMP-dependent protein kinase regulator
Length=402
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 53/178 (29%), Positives = 88/178 (49%), Gaps = 27/178 (15%)
Query 11 TAPTSVVQLTEDDLDSNEDDDDVVDDLPMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPF 70
T +S V + E+D +S+ D +D ++P+PS + R SV AE + ++ P
Sbjct 60 TQESSAVPVIEEDGESDSDSEDADLEVPVPS---KFTRRVSVCAETFNPDEEEEDNDPRV 116
Query 71 -YPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYL 129
+PKT+E++ R+++ ++ L +L+ E L +L A E V D +I+QG DGD Y+
Sbjct 117 VHPKTDEQRCRLQEACKDILLFKNLDQEQLSQVLDAMFEKIVKTDEHVIDQGDDGDNFYV 176
Query 130 IEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
IE G ++ ++ Q +SV Q + + FGELALMY
Sbjct 177 IERGTYDIL--VTKDNQ----TRSVGQYDNRGS-----------------FGELALMY 211
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 17/112 (15%)
Query 76 EEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDT-TIIEQGADGDKLYLIEEGE 134
++++ +E+ L SLE + ++ E K+ KD II QG D Y+IE GE
Sbjct 245 KKRKMFESFIESVPLFKSLEMSERMKIVDVIGE-KIYKDGERIIAQGEKADSFYIIESGE 303
Query 135 ANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
++ ++ + +++ Q+ + G+ FGELAL+
Sbjct 304 VSIL---------------IRSKTKSNKNGGNQEVEIAHCHKGQYFGELALV 340
> tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit;
K04739 cAMP-dependent protein kinase regulator
Length=285
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 63/123 (51%), Gaps = 28/123 (22%)
Query 71 YPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAF--KELKVPKDTT----IIEQGADG 124
Y KT +E+ +I +++++ FL S + + L++L+ AF K K+ ++ +I+QG DG
Sbjct 13 YEKTPQEQAKIMEMIKSCFLFSGVTSSGLDLLVKAFDFKAAKIKLFSSAGDVLIKQGDDG 72
Query 125 DKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELA 184
DKLYLIE G V KK + ++ + + G+ FGELA
Sbjct 73 DKLYLIESGTVEVTKKSASG----------------------AEEFLCTLTDGDYFGELA 110
Query 185 LMY 187
LMY
Sbjct 111 LMY 113
> hsa:5576 PRKAR2A, MGC3606, PKR2, PRKAR2; protein kinase, cAMP-dependent,
regulatory, type II, alpha (EC:2.7.11.1); K04739
cAMP-dependent protein kinase regulator
Length=404
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 45/152 (29%), Positives = 71/152 (46%), Gaps = 27/152 (17%)
Query 37 LPMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPP-FYPKTEEEKQRIRDIVENSFLISSLE 95
+P+PS + R SV AE Y ++ P +PKT+E++ R+++ ++ L +L+
Sbjct 88 VPVPS---RFNRRVSVCAETYNPDEEEEDTDPRVIHPKTDEQRCRLQEACKDILLFKNLD 144
Query 96 AEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQ 155
E L +L A E V D +I+QG DGD Y+IE G ++ + +SV
Sbjct 145 QEQLSQVLDAMFERIVKADEHVIDQGDDGDNFYVIERGTYDIL------VTKDNQTRSVG 198
Query 156 QNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
Q + + FGELALMY
Sbjct 199 QYDNRGS-----------------FGELALMY 213
Score = 35.0 bits (79), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 51/112 (45%), Gaps = 17/112 (15%)
Query 76 EEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDT-TIIEQGADGDKLYLIEEGE 134
++++ +E+ L+ SLE + ++ E K+ KD II QG D Y+IE GE
Sbjct 247 KKRKMFESFIESVPLLKSLEVSERMKIVDVIGE-KIYKDGERIITQGEKADSFYIIESGE 305
Query 135 ANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
++ ++ + ++D Q+ + G+ FGELAL+
Sbjct 306 VSIL---------------IRSRTKSNKDGGNQEVEIARCHKGQYFGELALV 342
> dre:100329437 protein kinase, cAMP-dependent, regulatory, type
II, alpha-like
Length=385
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 69/155 (44%), Gaps = 31/155 (20%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHF-----VPPFYPKTEEEKQRIRDIVENSFLIS 92
P PS + R SV AE Y + + KT+E++QR++D ++ L
Sbjct 107 PPPS---KCNRRVSVCAEPYNPDEDDDDGDEGSQLTVLHQKTDEQRQRLQDACKHILLFK 163
Query 93 SLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKK 152
+LE + L +L + E+ V II QG DGD Y+IE G VY+ + QQ +
Sbjct 164 TLEEDQLAEVLDSMFEVLVKPGECIINQGDDGDNFYVIERG---VYEIVI---QQDGLQH 217
Query 153 SVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
SV + + + FGELALMY
Sbjct 218 SVGRYDHKGS-----------------FGELALMY 235
> dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kinase,
cAMP-dependent, regulatory, type II, alpha, B; K04739
cAMP-dependent protein kinase regulator
Length=422
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 69/155 (44%), Gaps = 31/155 (20%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHF-----VPPFYPKTEEEKQRIRDIVENSFLIS 92
P PS + R SV AE Y + + KT+E++QR++D ++ L
Sbjct 107 PPPS---KCNRRVSVCAEPYNPDEDDDDGDEGSQLTVLHQKTDEQRQRLQDACKHILLFK 163
Query 93 SLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKK 152
+LE + L +L + E+ V II QG DGD Y+IE G VY+ + QQ +
Sbjct 164 TLEEDQLAEVLDSMFEVLVKPGECIINQGDDGDNFYVIERG---VYEIVI---QQDGLQH 217
Query 153 SVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
SV + + + FGELALMY
Sbjct 218 SVGRYDHKGS-----------------FGELALMY 235
> mmu:19088 Prkar2b, AI451071, AW061005, Pkarb2, RII(beta); protein
kinase, cAMP dependent regulatory, type II beta (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=416
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 66/143 (46%), Gaps = 30/143 (20%)
Query 49 RQSVSAEVYG----EWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLG 104
R SV AE Y E + ++ + +PKT++++ R+++ ++ L +L+ E + +L
Sbjct 110 RASVCAEAYNPDEEEDDAESRII---HPKTDDQRNRLQEACKDILLFKNLDPEQMSQVLD 166
Query 105 AFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDK 164
A E V + +I+QG DGD Y+I+ G ++Y K + V + +
Sbjct 167 AMFEKLVKEGEHVIDQGDDGDNFYVIDRGTFDIYVKCDGV------GRCVGNYDNRGS-- 218
Query 165 QQQQQLVNVMVAGELFGELALMY 187
FGELALMY
Sbjct 219 ---------------FGELALMY 226
> hsa:5577 PRKAR2B, PRKAR2, RII-BETA; protein kinase, cAMP-dependent,
regulatory, type II, beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=418
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 65/143 (45%), Gaps = 30/143 (20%)
Query 49 RQSVSAEVYG----EWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLG 104
R SV AE Y E + ++ + +PKT++++ R+++ ++ L +L+ E + +L
Sbjct 112 RASVCAEAYNPDEEEDDAESRII---HPKTDDQRNRLQEACKDILLFKNLDPEQMSQVLD 168
Query 105 AFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDK 164
A E V +I+QG DGD Y+I+ G ++Y K + V + +
Sbjct 169 AMFEKLVKDGEHVIDQGDDGDNFYVIDRGTFDIYVKCDGV------GRCVGNYDNRGS-- 220
Query 165 QQQQQLVNVMVAGELFGELALMY 187
FGELALMY
Sbjct 221 ---------------FGELALMY 228
> cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=780
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 64/149 (42%), Gaps = 29/149 (19%)
Query 39 MPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAED 98
+P+ VQ + +VSAE NK Y KT KQ IRD V+ + + L E
Sbjct 152 LPADGVQRAKKIAVSAEPTNFENKPATL--QHYNKTVGAKQMIRDAVQKNDFLKQLAKEQ 209
Query 99 LEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNE 158
+ L+ E++ +I++G GD+L+++ EGE V
Sbjct 210 IIELVNCMYEMRARAGQWVIQEGEPGDRLFVVAEGELQV--------------------- 248
Query 159 QQSEDKQQQQQLVNVMVAGELFGELALMY 187
++ L+ M AG + GELA++Y
Sbjct 249 ------SREGALLGKMRAGTVMGELAILY 271
Score = 29.6 bits (65), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 31/70 (44%), Gaps = 21/70 (30%)
Query 117 IIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVA 176
II QG GD ++I G+ V QQ E ++E ++ + V+
Sbjct 346 IIRQGEKGDAFFVINSGQVKV----------------TQQIEGETEPRE-----IRVLNQ 384
Query 177 GELFGELALM 186
G+ FGE AL+
Sbjct 385 GDFFGERALL 394
> dre:100331387 cAMP-dependent protein kinase, regulatory subunit
beta 2-like; K04739 cAMP-dependent protein kinase regulator
Length=403
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 41/140 (29%), Positives = 62/140 (44%), Gaps = 24/140 (17%)
Query 49 RQSVSAEVYGEWNKKTHFVPPF-YPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFK 107
R SV AE Y + P YPKT+E++QR+++ ++ L +L+ E + +L A
Sbjct 97 RASVCAEAYNPDEDEEEREPRVTYPKTDEQRQRLQEACKDILLFKNLDQEQMSQVLDAMF 156
Query 108 ELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQ 167
E V II+Q DGD Y+IE G + K + ++V + +
Sbjct 157 EKVVVSGEHIIDQDDDGDNFYVIERGMFEILLKADGT------TRTVGSYDNRGS----- 205
Query 168 QQLVNVMVAGELFGELALMY 187
FGELALMY
Sbjct 206 ------------FGELALMY 213
> xla:414594 prkar2b, MGC83177; protein kinase, cAMP-dependent,
regulatory, type II, beta; K04739 cAMP-dependent protein kinase
regulator
Length=402
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 51/94 (54%), Gaps = 3/94 (3%)
Query 49 RQSVSAEVYG--EWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAF 106
R SV AE Y E + T V +PKT++++ R+++ + L +L+ E + +L A
Sbjct 96 RASVCAEAYNPDEEDDDTE-VKILFPKTDDQRNRLQEACTDILLFKNLDQEQMSQVLDAM 154
Query 107 KELKVPKDTTIIEQGADGDKLYLIEEGEANVYKK 140
E V +I+QG DGD Y+I+ G +++ K
Sbjct 155 FEKLVQCGEHVIDQGDDGDNFYVIDRGTYDIFVK 188
> dre:556042 prkar2aa, MGC153742, wu:fb55f12, wu:fj29e12, wu:fj33e06,
zgc:153742; protein kinase, cAMP-dependent, regulatory,
type II, alpha A; K04739 cAMP-dependent protein kinase regulator
Length=397
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 68/151 (45%), Gaps = 27/151 (17%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPF-YPKTEEEKQRIRDIVENSFLISSLEA 96
P PS + R SV AE + + + P +PKT+E++ R+++ ++ L +L+
Sbjct 86 PPPS---RFNRRVSVCAEAFNPDDDEEDSEPRVVHPKTDEQRCRLQEACKDILLFKALDQ 142
Query 97 EDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQ 156
E +L A E+ V +I+QG DGD Y+IE G VY + Q+ Q
Sbjct 143 EQFSEVLDAMFEVLVQPQDHVIDQGDDGDNFYVIERG---VYDIV--VQKDGVGCCVGQY 197
Query 157 NEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
N + S FGELALMY
Sbjct 198 NNKGS------------------FGELALMY 210
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 49/115 (42%), Gaps = 17/115 (14%)
Query 73 KTEEEKQRIRDI-VENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIE 131
K +K+R+ + +E+ L+ SLE + ++ II QG D Y++E
Sbjct 240 KNNAKKRRMYECFIESIPLLKSLELSERMKIVDVLGVKTFQDGERIIMQGDKADCFYIVE 299
Query 132 EGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
GE + K S ++ +QD V+ + G+ FGELAL+
Sbjct 300 SGEVKIMMK-SKTKADRQDNAEVE---------------IARCSRGQYFGELALV 338
> hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR1,
PPNAD1, PRKAR1, TSE1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 54/102 (52%), Gaps = 2/102 (1%)
Query 38 PMPSAKVQAKNRQ-SVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEA 96
P P+ V+ + R+ ++SAEVY E + + +V PK + + +E + L S L+
Sbjct 85 PPPNPVVKGRRRRGAISAEVYTEEDAAS-YVRKVIPKDYKTMAALAKAIEKNVLFSHLDD 143
Query 97 EDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ + A + T+I+QG +GD Y+I++GE +VY
Sbjct 144 NERSDIFDAMFSVSFIAGETVIQQGDEGDNFYVIDQGETDVY 185
> mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protein
kinase, cAMP dependent regulatory, type I, alpha (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 62/124 (50%), Gaps = 6/124 (4%)
Query 16 VVQLTEDDLDSNEDDDDVVDDLPMPSAKVQAKNRQ-SVSAEVYGEWNKKTHFVPPFYPKT 74
+Q T DS ED+ P P+ V+ + R+ ++SAEVY E + + +V PK
Sbjct 67 CLQKTGIRTDSREDEISP----PPPNPVVKGRRRRGAISAEVYTEEDAAS-YVRKVIPKD 121
Query 75 EEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGE 134
+ + +E + L S L+ + + A + T+I+QG +GD Y+I++GE
Sbjct 122 YKTMAALAKAIEKNVLFSHLDDNERSDIFDAMFPVSFIAGETVIQQGDEGDNFYVIDQGE 181
Query 135 ANVY 138
+VY
Sbjct 182 MDVY 185
> ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regulator/
catalytic/ protein kinase/ protein serine/threonine
phosphatase; K04345 protein kinase A [EC:2.7.11.11]
Length=1094
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 58/124 (46%), Gaps = 19/124 (15%)
Query 66 FVPP--FYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGAD 123
+VPP + KT EE+ I ++ + FL L +VLL + L+ +++QG +
Sbjct 465 WVPPSPAHRKTWEEEAHIERVLRDHFLFRKLTDSQCQVLLDCMQRLEANPGDIVVKQGGE 524
Query 124 GDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGEL 183
GD Y++ GE V + Q ++ + + ++ + +KQ FGEL
Sbjct 525 GDCFYVVGSGEFEVL-----ATQDGKNGEVPRILQRYTAEKQSS------------FGEL 567
Query 184 ALMY 187
ALM+
Sbjct 568 ALMH 571
> dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein
kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific
extinguisher 1) a
Length=379
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 49/101 (48%), Gaps = 1/101 (0%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAE 97
PM + R ++SAEVY E T +V PK + + +E + L + L+
Sbjct 84 PMNPVVKGRRRRGAISAEVYTE-EDATSYVRKVIPKDYKTMAALAKAIEKNVLFAHLDDN 142
Query 98 DLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ + A + T+I+QG +GD Y+I++GE +VY
Sbjct 143 ERSDIFDAMFSVTYIAGETVIQQGDEGDNFYVIDQGEMDVY 183
> dre:100329979 protein kinase, cAMP-dependent, regulatory, type
I, alpha (tissue specific extinguisher 1) a-like; K04739
cAMP-dependent protein kinase regulator
Length=379
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 49/101 (48%), Gaps = 1/101 (0%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAE 97
PM + R ++SAEVY E T +V PK + + +E + L + L+
Sbjct 84 PMNPVVKGRRRRGAISAEVYTE-EDATSYVRKVIPKDYKTMAALAKAIEKNVLFAHLDDN 142
Query 98 DLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ + A + T+I+QG +GD Y+I++GE +VY
Sbjct 143 ERSDIFDAMFSVTYIAGETVIQQGDEGDNFYVIDQGEMDVY 183
> xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dependent,
regulatory, type I,beta; K04739 cAMP-dependent protein
kinase regulator
Length=381
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 60/115 (52%), Gaps = 5/115 (4%)
Query 25 DSNEDDDDVVDDLPMPSAKVQAKNRQ-SVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRD 83
DS ED+ + P ++ V+ + R+ ++SAEVY E + + +V PK + +
Sbjct 75 DSREDE---ISPPPHMNSVVKGRRRRGAISAEVYTEEDAAS-YVRKVIPKDYKTMAALAK 130
Query 84 IVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+E + L + L+ + + A + T+I+QG +GD Y++++GE +VY
Sbjct 131 AIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNFYVVDQGEMDVY 185
> xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 60/115 (52%), Gaps = 5/115 (4%)
Query 25 DSNEDDDDVVDDLPMPSAKVQAKNRQ-SVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRD 83
DS ED+ + P ++ V+ + R+ ++SAEVY E + + +V PK + +
Sbjct 75 DSREDE---ISPPPHMNSVVKGRRRRGAISAEVYTEEDAAS-YVRKVIPKDYKTMAALAK 130
Query 84 IVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+E + L + L+ + + A + T+I+QG +GD Y++++GE +VY
Sbjct 131 AIEKNVLFAHLDDTERSDIFDAMFSVTYISGETVIQQGDEGDNFYVVDQGEMDVY 185
> mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP dependent
regulatory, type I beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 53/113 (46%), Gaps = 2/113 (1%)
Query 26 SNEDDDDVVDDLPMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIV 85
+ D+++ P P K + + R VSAEVY E +V PK + + +
Sbjct 75 CDSHDEEISPTPPNPVVKAR-RRRGGVSAEVYTE-EDAVSYVRKVIPKDYKTMTALAKAI 132
Query 86 ENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ L S L+ + + A + T+I+QG +GD Y+I++GE +VY
Sbjct 133 SKNVLFSHLDDNERSDIFDAMFPVTHIGGETVIQQGNEGDNFYVIDQGEVDVY 185
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 117 IIEQGADGDKLYLIEEGEANVYKKISPSQQ 146
I+ QG GD Y+I EG A+V ++ SP+++
Sbjct 282 IVVQGEPGDDFYIITEGTASVLQRRSPNEE 311
> hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regulatory,
type I, beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 54/113 (47%), Gaps = 2/113 (1%)
Query 26 SNEDDDDVVDDLPMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIV 85
S+ D++V P P K + + R VSAEVY E +V PK + + +
Sbjct 75 SDSHDEEVSPTPPNPVVKAR-RRRGGVSAEVYTE-EDAVSYVRKVIPKDYKTMTALAKAI 132
Query 86 ENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ L + L+ + + A + T+I+QG +GD Y++++GE +VY
Sbjct 133 SKNVLFAHLDDNERSDIFDAMFPVTHIAGETVIQQGNEGDNFYVVDQGEVDVY 185
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 117 IIEQGADGDKLYLIEEGEANVYKKISPSQQ 146
I+ QG GD Y+I EG A+V ++ SP+++
Sbjct 282 IVVQGEPGDDFYIITEGTASVLQRRSPNEE 311
> dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependent,
regulatory, type I, alpha (tissue specific extinguisher
1) b (EC:2.7.11.1); K04739 cAMP-dependent protein kinase regulator
Length=379
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 49/101 (48%), Gaps = 1/101 (0%)
Query 38 PMPSAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAE 97
PM + R ++SAEVY E + + +V PK + + +E + L S L+
Sbjct 84 PMNPVVKGRRRRGAISAEVYTEEDAAS-YVRKVIPKDYKTMAALAKAIEKNVLFSHLDDN 142
Query 98 DLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ + A + +I+QG +GD Y+I++GE +VY
Sbjct 143 ERSDIFDAMFPVTYIAGEIVIQQGDEGDNFYVIDQGEMDVY 183
> bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:2.7.11.1);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=887
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/115 (21%), Positives = 50/115 (43%), Gaps = 27/115 (23%)
Query 73 KTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEE 132
KT+++ IR + N+ + SL +++ +G+ ++P ++ QG +G ++I E
Sbjct 64 KTDKDITLIRHALANNLVCESLNEYEIDAFIGSMSSFELPAGAPVVRQGDNGTYFFIISE 123
Query 133 GEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
G+ +VY + VN M G FGE++L++
Sbjct 124 GDFDVY---------------------------VDGEHVNSMTRGTAFGEISLIH 151
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 47/98 (47%), Gaps = 2/98 (2%)
Query 41 SAKVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLE 100
SA V+ KN+ +++G + F + R+ ++ + +L E+
Sbjct 156 SATVKVKNKAGNCGKLWGV--TRVVFRDTLKRISLRNYTENREFIDCVTIFENLTEENKS 213
Query 101 VLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVY 138
+ A EL+ +I++QG GD LYLI++G A+VY
Sbjct 214 CITNALVELRFSPGESIVKQGDPGDDLYLIKQGSADVY 251
Score = 36.6 bits (83), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 89 FLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYK 139
+++ L + +++L+ A K L+ + TI +G GD LY+I+ GE + K
Sbjct 449 YILRHLSEKQIDMLIKALKTLRYKLNDTIFNEGEIGDMLYIIKSGEVAIIK 499
> dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAMP-dependent,
regulatory, type I, beta (EC:2.7.11.1); K04739
cAMP-dependent protein kinase regulator
Length=380
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/141 (24%), Positives = 56/141 (39%), Gaps = 28/141 (19%)
Query 47 KNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAF 106
+ R VSAEVY E +V PK + + + + L + L+ + + A
Sbjct 94 RRRGGVSAEVYTE-EDAVSYVRKVIPKDYKTMTALAKAISKNVLFAHLDDNERSDIFDAM 152
Query 107 KELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQ 166
+ T+I+QG +GD Y+I++GE +VY
Sbjct 153 FPVTHIAGETVIQQGDEGDNFYVIDQGEVDVY---------------------------V 185
Query 167 QQQLVNVMVAGELFGELALMY 187
+ V + G FGELAL+Y
Sbjct 186 NGEWVTSIGEGGSFGELALIY 206
> cel:R07E4.6 kin-2; protein KINase family member (kin-2); K04739
cAMP-dependent protein kinase regulator
Length=376
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 35/145 (24%), Positives = 58/145 (40%), Gaps = 29/145 (20%)
Query 43 KVQAKNRQSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVL 102
K R +SAE E T + PK + ++ + + + L + LE ++ + +
Sbjct 87 KRSGGRRTGISAEPIKE--DDTEYKKVVIPKDDATRRSLESAMRKNLLFAHLEEDEQKTM 144
Query 103 LGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSE 162
A ++ TIIEQG +GD Y+I++G +VY
Sbjct 145 YDAMFPVEKSAGETIIEQGEEGDNFYVIDKGTVDVY------------------------ 180
Query 163 DKQQQQQLVNVMVAGELFGELALMY 187
+ V + G FGELAL+Y
Sbjct 181 ---VNHEYVLTINEGGSFGELALIY 202
Score = 34.3 bits (77), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 115 TTIIEQGADGDKLYLIEEGEANVYKKIS 142
T ++EQG GD+ ++I EGEANV +K S
Sbjct 275 THVVEQGQPGDEFFIILEGEANVLQKRS 302
> cel:C09G4.2 hypothetical protein
Length=617
Score = 40.0 bits (92), Expect = 0.004, Method: Composition-based stats.
Identities = 30/101 (29%), Positives = 44/101 (43%), Gaps = 28/101 (27%)
Query 87 NSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQ 146
NSFL +L+A +E + A ++VP II QG G +Y+I+EG+ V K
Sbjct 70 NSFL-RNLDATQIEKISSAMYPVEVPAGAIIIRQGDLGSIMYVIQEGKVQVVK------- 121
Query 147 QQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ V M G LFGELA+++
Sbjct 122 --------------------DNRFVRTMEDGALFGELAILH 142
> dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5
Length=663
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/114 (22%), Positives = 56/114 (49%), Gaps = 23/114 (20%)
Query 73 KTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEE 132
K++ +++++ ++ + + +L L ++ + +E++ + I+ +GA+G+ Y+I +
Sbjct 208 KSKMKREQLMGFLKTARTLKALNDVQLSKIIDSMEEVRFQDNEVIVREGAEGNTFYIILK 267
Query 133 GEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
GE V KK++ QQ+ + M GE FGELAL+
Sbjct 268 GEVLVTKKVNG-----------------------QQKTIRKMGEGEHFGELALI 298
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 50/114 (43%), Gaps = 28/114 (24%)
Query 74 TEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEG 133
+ E Q ++ I +N FL L+ E + +++ K L II++G +GD +Y++ G
Sbjct 92 SSETSQIVKAIGKNDFL-RRLDEEQISMMVDLMKVLDCRAGDEIIKEGTEGDSMYIVAAG 150
Query 134 EANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
E V Q D + ++ G++FGELA++Y
Sbjct 151 ELKV--------------------TQAGRD-------LRILSPGDMFGELAILY 177
> mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b;
protein kinase, cGMP-dependent, type I (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 73/156 (46%), Gaps = 34/156 (21%)
Query 37 LPMPSAKV--QAKNRQSVSAE--VYGEWNKKTHFVPPFYPKTEEEKQRIRD-IVENSFLI 91
LP+PS + + Q +SAE Y ++ F K+E K I++ I++N F+
Sbjct 47 LPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKF-TKSERSKDLIKEAILDNDFM- 104
Query 92 SSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDK 151
+LE ++ ++ ++ KD+ II++G G +Y++E+G+ V K
Sbjct 105 KNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEDGKVEV------------TK 152
Query 152 KSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ V+ + M G++FGELA++Y
Sbjct 153 EGVK---------------LCTMGPGKVFGELAILY 173
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 93 SLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQ 145
SL E L L +E II QGA GD ++I +G+ NV ++ SPS+
Sbjct 224 SLPDEILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGQVNVTREDSPSE 276
> hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK,
PKG, PRKG1B, PRKGR1B, cGKI-BETA, cGKI-alpha; protein kinase,
cGMP-dependent, type I (EC:2.7.11.12); K07376 protein kinase,
cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 73/156 (46%), Gaps = 34/156 (21%)
Query 37 LPMPSAKV--QAKNRQSVSAE--VYGEWNKKTHFVPPFYPKTEEEKQRIRD-IVENSFLI 91
LP+PS + + Q +SAE Y ++ F K+E K I++ I++N F+
Sbjct 47 LPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKF-TKSERSKDLIKEAILDNDFM- 104
Query 92 SSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDK 151
+LE ++ ++ ++ KD+ II++G G +Y++E+G+ V K
Sbjct 105 KNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEDGKVEV------------TK 152
Query 152 KSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
+ V+ + M G++FGELA++Y
Sbjct 153 EGVK---------------LCTMGPGKVFGELAILY 173
Score = 34.7 bits (78), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 93 SLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQ 145
SL E L L +E II QGA GD ++I +G NV ++ SPS+
Sbjct 224 SLPEEILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGTVNVTREDSPSE 276
> dre:100125912 prkg2; protein kinase, cGMP-dependent, type II
(EC:2.7.11.1); K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=768
Score = 36.2 bits (82), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 26/112 (23%), Positives = 49/112 (43%), Gaps = 22/112 (19%)
Query 75 EEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGE 134
E ++ R+ + + L++SL + L ++ +E K II +G +G+ Y+I G+
Sbjct 278 EARHEQYRNFLRSVSLLASLPGDKLTKIVDCLEEY-YNKGDYIIREGEEGNTFYIIANGK 336
Query 135 ANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
V QS ++ Q++N + G+ FGE AL+
Sbjct 337 IKV---------------------TQSTQDHEEPQIINTLGKGDYFGEKALI 367
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 30/148 (20%), Positives = 61/148 (41%), Gaps = 36/148 (24%)
Query 49 RQSVSAEVYGEWNKKTH---FVPPFYP------KTEEEKQRIRDIVENSFLISSLEAEDL 99
R+ A V E + +T+ +P FY K + K+ + D + + + LE + +
Sbjct 125 RKGAKAGVSAEPSSRTYDSSGIPKFYFESARVWKEQSVKKLLTDALNKNQYLRRLEVQQV 184
Query 100 EVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQ 159
+ ++ E + +I+QG G+ L+++ +G+ +VY
Sbjct 185 KDMVECMYERTYQQGEYVIKQGEPGNHLFVLADGKLDVY--------------------- 223
Query 160 QSEDKQQQQQLVNVMVAGELFGELALMY 187
Q +L+ + FGELA++Y
Sbjct 224 ------QHNKLLTSIAVWTTFGELAILY 245
> cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase
with 3 cNMP binding domains and a Ser/Thr kinase domain ;
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=965
Score = 35.8 bits (81), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 77 EKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEAN 136
E R RD +++ F+ + + L +L+ + + +K I+ QG G ++++ GE
Sbjct 486 EINRKRDAIKSCFVFQYVSEQQLSLLVKSLRLVKFTSGEKIVVQGDKGTAFFILQSGEVA 545
Query 137 VYK 139
VY+
Sbjct 546 VYR 548
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 45/115 (39%), Gaps = 27/115 (23%)
Query 73 KTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEE 132
KTEE+ + I + + + +SL ++ L+ + + +IEQGA G ++I
Sbjct 92 KTEEDIKTISKALAGNVVGASLNESEIATLVSSMHYYEYEVGEVVIEQGASGFYFFVIST 151
Query 133 GEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
G V + N VN M G FGELAL++
Sbjct 152 GSFGV---------------EINGNR------------VNTMSEGTAFGELALIH 179
> tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=994
Score = 35.8 bits (81), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 30/58 (51%), Gaps = 0/58 (0%)
Query 82 RDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYK 139
R I+ ++ L + +L+ AFK ++ II++G G + ++I+ GE + K
Sbjct 553 RAIIRKMYIFRYLSDHQMTMLIKAFKTVRYMSGEYIIKEGERGTRFFIIKAGEVAILK 610
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 47/115 (40%), Gaps = 27/115 (23%)
Query 73 KTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEE 132
KT + I+D ++ + + SSL +++ L A + K + +QG G ++I
Sbjct 172 KTPADFALIQDSLKANLVCSSLNEGEIDALAVAMQFFTFKKGDVVTKQGEPGSYFFIIHS 231
Query 133 GEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
G +V DK+ VN M G+ FGE+AL++
Sbjct 232 GTFDVL---------VNDKR------------------VNAMDKGKAFGEIALIH 259
> tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=892
Score = 33.9 bits (76), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 25/114 (21%), Positives = 46/114 (40%), Gaps = 27/114 (23%)
Query 73 KTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEE 132
K+E +K I+ + N+ + S+L ++ + + + + EQG +G ++I E
Sbjct 63 KSESDKSFIKKSLANNVIFSALNDLEMSAFVDSMSYYVFSVGSKVTEQGTNGSYFFVINE 122
Query 133 GEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
G +VY +LVN M G FGE++L+
Sbjct 123 GIFDVY---------------------------IDGKLVNTMERGTAFGEISLI 149
Score = 32.7 bits (73), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 38/138 (27%), Positives = 56/138 (40%), Gaps = 48/138 (34%)
Query 50 QSVSAEVYGEWNKKTHFVPPFYPKTEEEKQRIRDIVENSFLISSLEAEDLEVLLGAFKEL 109
+++S EVY + V F TE +K ++V N+F+ S
Sbjct 190 KNISMEVYSQNRSFLDSVKIFEMLTENQK----NMVTNAFVESKF--------------- 230
Query 110 KVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQ 169
VP D I+ QG GD LY+I+EG+A+V D+K
Sbjct 231 -VPGDR-IVTQGDFGDVLYIIKEGKADVL---------INDEK----------------- 262
Query 170 LVNVMVAGELFGELALMY 187
V + G+ FGE AL+Y
Sbjct 263 -VRTLTNGQYFGERALLY 279
> pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=853
Score = 32.3 bits (72), Expect = 0.96, Method: Composition-based stats.
Identities = 14/58 (24%), Positives = 31/58 (53%), Gaps = 0/58 (0%)
Query 82 RDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYK 139
+ I++ ++ L + +L+ AF+ + + II++G G + Y+I+ GE + K
Sbjct 410 KSIIKKMYIFRYLTDKQCNLLIEAFRTTRYEEGDYIIQEGEVGSRFYIIKNGEVEIVK 467
Score = 31.6 bits (70), Expect = 1.8, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 34/72 (47%), Gaps = 27/72 (37%)
Query 116 TIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMV 175
TI++QG GD LY+++EG+A VY N+++ + V+
Sbjct 202 TIVKQGDYGDVLYILKEGKATVYI-----------------NDEE----------IRVLE 234
Query 176 AGELFGELALMY 187
G FGE AL+Y
Sbjct 235 KGSYFGERALLY 246
> dre:566430 cGMP dependent protein kinase I
Length=528
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 93 SLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPS 144
L+ E L L +E II QGA GD ++I +G+ NV ++ +P+
Sbjct 81 GLQEEILSKLADVLEETHYSDGEYIIRQGARGDTFFIISKGKVNVTREDAPN 132
> mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II;
protein kinase, cGMP-dependent, type II (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=762
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 23/105 (21%), Positives = 43/105 (40%), Gaps = 21/105 (20%)
Query 82 RDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKI 141
R+ + + L+ +L + L ++ + K II +G +G +++ +G+ V
Sbjct 278 RNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKGKVKV---- 333
Query 142 SPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
QS + Q QL+ + GE FGE AL+
Sbjct 334 -----------------TQSTEGHDQPQLIKTLQKGEYFGEKALI 361
> hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,
type II (EC:2.7.11.12); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=762
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/105 (21%), Positives = 43/105 (40%), Gaps = 21/105 (20%)
Query 82 RDIVENSFLISSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKI 141
R+ + + L+ +L + L ++ + K II +G +G +++ +G+ V
Sbjct 278 RNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKGKVKV---- 333
Query 142 SPSQQQQQDKKSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALM 186
QS + Q QL+ + GE FGE AL+
Sbjct 334 -----------------TQSTEGHDQPQLIKTLQKGEYFGEKALI 361
Score = 29.6 bits (65), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 31/156 (19%), Positives = 63/156 (40%), Gaps = 36/156 (23%)
Query 41 SAKVQAKNRQSVSAEVYGEWNKKTHFV--PPFYP-------KTEEEKQRIRDIVENSFLI 91
S V +R+ A V E +T+ + PP + K EK+ I D + + +
Sbjct 110 SGLVSLHSRRGAKAGVSAEPTTRTYDLNKPPEFSFEKARVRKDSSEKKLITDALNKNQFL 169
Query 92 SSLEAEDLEVLLGAFKELKVPKDTTIIEQGADGDKLYLIEEGEANVYKKISPSQQQQQDK 151
L+ + ++ ++ + + II+QG G+ ++++ EG V+
Sbjct 170 KRLDPQQIKDMVECMYGRNYQQGSYIIKQGEPGNHIFVLAEGRLEVF------------- 216
Query 152 KSVQQNEQQSEDKQQQQQLVNVMVAGELFGELALMY 187
Q ++L++ + FGELA++Y
Sbjct 217 --------------QGEKLLSSIPMWTTFGELAILY 238
Lambda K H
0.307 0.126 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5235419772
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40