bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0094_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
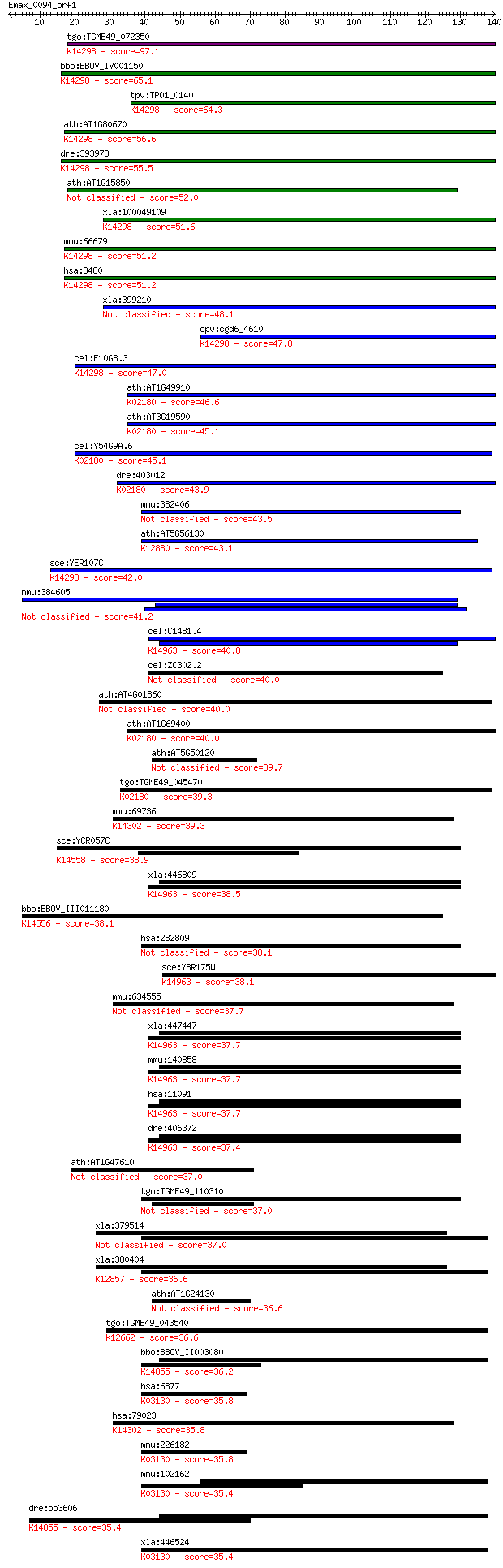
tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K142... 97.1 1e-20
bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA ... 65.1 7e-11
tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor 64.3 1e-10
ath:AT1G80670 transducin family protein / WD-40 repeat family ... 56.6 2e-08
dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA expo... 55.5 5e-08
ath:AT1G15850 transducin family protein / WD-40 repeat family ... 52.0 6e-07
xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA expo... 51.6 7e-07
mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41; R... 51.2 1e-06
hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG1... 51.2 1e-06
xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein 48.1 8e-06
cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor 47.8 1e-05
cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member... 47.0 2e-05
ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint... 46.6 2e-05
ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint... 45.1 6e-05
cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3); K... 45.1 7e-05
dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibit... 43.9 2e-04
mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar prote... 43.5 2e-04
ath:AT5G56130 transducin family protein / WD-40 repeat family ... 43.1 3e-04
sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of t... 42.0 5e-04
mmu:384605 Wdr88, Gm1429, Pqwd; WD repeat domain 88 41.2
cel:C14B1.4 tag-125; Temporarily Assigned Gene name family mem... 40.8 0.001
cel:ZC302.2 hypothetical protein 40.0 0.002
ath:AT4G01860 transducin family protein / WD-40 repeat family ... 40.0 0.002
ath:AT1G69400 transducin family protein / WD-40 repeat family ... 40.0 0.002
ath:AT5G50120 transducin family protein / WD-40 repeat family ... 39.7 0.003
tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative ; ... 39.3 0.004
mmu:69736 Nup37, 2410003L22Rik, 2810039M17Rik; nucleoporin 37;... 39.3 0.004
sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ri... 38.9 0.005
xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat dom... 38.5 0.007
bbo:BBOV_III011180 17.m07962; WD domain, G-beta repeat contain... 38.1 0.008
hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B; PO... 38.1 0.009
sce:YBR175W SWD3, CPS30, SAF35; Essential subunit of the COMPA... 38.1 0.009
mmu:634555 nucleoporin Nup37-like 37.7 0.011
xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repe... 37.7 0.012
mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3... 37.7 0.012
hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPAS... 37.7 0.012
dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat d... 37.4 0.014
ath:AT1G47610 transducin family protein / WD-40 repeat family ... 37.0 0.019
tgo:TGME49_110310 WD domain, G-beta repeat-containing protein 37.0 0.019
xla:379514 snrnp40-a, MGC64565, prp8bp, snrnp40, spf38, wdr57;... 37.0 0.020
xla:380404 snrnp40-b, MGC132119, MGC53216, prp8bp, spf38, wdr5... 36.6 0.023
ath:AT1G24130 transducin family protein / WD-40 repeat family ... 36.6 0.023
tgo:TGME49_043540 U4/U6 small nuclear ribonucleoprotein, putat... 36.6 0.025
bbo:BBOV_II003080 18.m06257; WD-repeat protein; K14855 ribosom... 36.2 0.034
hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA b... 35.8 0.039
hsa:79023 NUP37, FLJ22618, MGC5585, p37; nucleoporin 37kDa; K1... 35.8 0.043
mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase ... 35.8 0.043
mmu:102162 Taf5l, 1110005N04Rik, AI849020; TAF5-like RNA polym... 35.4 0.048
dre:553606 nle1, MGC110281, fc12b09, fc45f01, wu:fc12b09, wu:f... 35.4 0.051
xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/... 35.4 0.055
> tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K14298
mRNA export factor
Length=375
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/122 (41%), Positives = 68/122 (55%), Gaps = 7/122 (5%)
Query 18 GRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAF 77
G + +YY+K ++T LPNGP D++SQL WS +G L+C SWD T RVWQI+ A F
Sbjct 14 GGAVNNQMSYYNKASATNLPNGPRDTISQLGWSNEGSLLSCTSWDNTVRVWQIS---AGF 70
Query 78 GTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
G+ Q K AP+L G + L CDK VK YDL + +T QV+ Q
Sbjct 71 GSQIQAAAK----VCMDAQAPLLCSTFGPSPNHLFVGCCDKTVKLYDLNASSSTPQVVAQ 126
Query 138 HE 139
H+
Sbjct 127 HD 128
> bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA
export factor
Length=359
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 59/126 (46%), Gaps = 15/126 (11%)
Query 16 AFGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQAL--ACASWDRTCRVWQITTQ 73
A+ RS+T+ + L P DS+S + WS L + SWD+T R+W+I+
Sbjct 2 AYSRSSTDDPKTHF------LTGIPDDSISHIRWSHSSNPLLLSAGSWDKTVRLWRISPN 55
Query 74 PAAFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQ 133
T+ V+ L+ + AP+L+ C D GC V AYDL S TG
Sbjct 56 IGNTLTSDCVV-------LYRQEAPILTSCFSDDNTKFFAGGCSNTVMAYDLASRNATGV 108
Query 134 VIGQHE 139
++ +H+
Sbjct 109 LVARHD 114
> tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor
Length=359
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 53/106 (50%), Gaps = 9/106 (8%)
Query 36 LPNGPSDSVSQLAWSR--DGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLF 93
L N P+DS+S L WS + L SWD+T R+W++TT N ++Y F
Sbjct 17 LNNLPNDSISHLRWSTTTNPLLLTAGSWDKTLRIWKVTTGLGN-AVNTDMVYT------F 69
Query 94 TETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
+ APVL + L GC V AYDL + +TG VI +H+
Sbjct 70 KQDAPVLCSAFSTDSMRLFGGGCTNNVLAYDLNNPSSTGVVIARHQ 115
> ath:AT1G80670 transducin family protein / WD-40 repeat family
protein; K14298 mRNA export factor
Length=349
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/125 (30%), Positives = 57/125 (45%), Gaps = 16/125 (12%)
Query 17 FGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAA 76
FG T ++N S E+ P+DS+S L++S L SWD R W+I+ A+
Sbjct 4 FGAPATANSN---PNKSYEVTPSPADSISSLSFSPRADILVATSWDNQVRCWEISRSGAS 60
Query 77 FGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQ--V 134
+ P++ + PVL D + + GCDKQ K + L+SG GQ
Sbjct 61 LAS--------APKASISHDQPVLCSAWKDDGTTVFSGGCDKQAKMWPLLSG---GQPVT 109
Query 135 IGQHE 139
+ HE
Sbjct 110 VAMHE 114
> dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 42/127 (33%), Positives = 57/127 (44%), Gaps = 19/127 (14%)
Query 16 AFGRSTTESANYYSKQNSTELPNGPSDSVSQLAWS---RDGQALACASWDRTCRVWQITT 72
FG +TT+S N E+ + P +S+S LA+S G L SW R W++
Sbjct 17 VFGSTTTDSHN---PMKDVEVTSPPDESISCLAFSPPTMPGNFLIGGSWANDVRCWEVQD 73
Query 73 QPAAFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTG 132
NGQ + P++ T PVL C D + TA CDK K +DL S +
Sbjct 74 -------NGQTV----PKAQQMHTGPVLDVCWSDDGSKVFTASCDKTAKMWDLNSNQAIQ 122
Query 133 QVIGQHE 139
I QHE
Sbjct 123 --IAQHE 127
> ath:AT1G15850 transducin family protein / WD-40 repeat family
protein
Length=140
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 49/111 (44%), Gaps = 10/111 (9%)
Query 18 GRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAF 77
G T S N + NS E+ +DS+S L++S L SWD R W+IT +
Sbjct 5 GDKATSSTN--NPNNSYEITPPATDSISSLSFSPKADILVATSWDCQVRCWEITRSDGSI 62
Query 78 GTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISG 128
+ P+ + PVL D + T GCDKQ K + L+SG
Sbjct 63 AS--------EPKVSMSHDQPVLCSAWKDDGTTVFTGGCDKQAKMWPLLSG 105
> xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA export
1 homolog; K14298 mRNA export factor
Length=368
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 52/115 (45%), Gaps = 16/115 (13%)
Query 28 YSKQNSTELPNGPSDSVSQLAWSRD---GQALACASWDRTCRVWQITTQPAAFGTNGQVL 84
++ E+ + P DS+S L++S G L SW R W++ NGQ +
Sbjct 26 HNPMKDIEVASPPDDSISCLSFSPPTLPGNFLIAGSWANDVRCWEVQD-------NGQTI 78
Query 85 YKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
P++ T PVL C D + TA CDK K +DL S ++ + QHE
Sbjct 79 ----PKAQQMHTGPVLDVCWSDDGTKVFTASCDKTAKMWDLNSNQSIQ--VAQHE 127
> mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41;
RAE1 RNA export 1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 56/126 (44%), Gaps = 19/126 (15%)
Query 17 FGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRD---GQALACASWDRTCRVWQITTQ 73
FG +TT++ N E+ + P DS+ L++S G L SW R W++
Sbjct 18 FGSTTTDNHN---PMKDIEVTSSPDDSIGCLSFSPPTLPGNFLIAGSWANDVRCWEVQD- 73
Query 74 PAAFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQ 133
+GQ + P++ T PVL C D + TA CDK K +DL S +
Sbjct 74 ------SGQTI----PKAQQMHTGPVLDVCWSDDGSKVFTASCDKTAKMWDLNSNQAIQ- 122
Query 134 VIGQHE 139
I QH+
Sbjct 123 -IAQHD 127
> hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG14,
MRNP41, Mnrp41, dJ481F12.3, dJ800J21.1; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 56/126 (44%), Gaps = 19/126 (15%)
Query 17 FGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRD---GQALACASWDRTCRVWQITTQ 73
FG +TT++ N E+ + P DS+ L++S G L SW R W++
Sbjct 18 FGSATTDNHN---PMKDIEVTSSPDDSIGCLSFSPPTLPGNFLIAGSWANDVRCWEVQD- 73
Query 74 PAAFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQ 133
+GQ + P++ T PVL C D + TA CDK K +DL S +
Sbjct 74 ------SGQTI----PKAQQMHTGPVLDVCWSDDGSKVFTASCDKTAKMWDLSSNQAIQ- 122
Query 134 VIGQHE 139
I QH+
Sbjct 123 -IAQHD 127
> xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein
Length=368
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 51/115 (44%), Gaps = 16/115 (13%)
Query 28 YSKQNSTELPNGPSDSVSQLAWSRD---GQALACASWDRTCRVWQITTQPAAFGTNGQVL 84
++ E+ + P DS+S L++S G L SW R W++ NGQ +
Sbjct 26 HNPMKDIEVASPPDDSISCLSFSPQTLPGNFLIAGSWANDVRCWEVQD-------NGQTI 78
Query 85 YKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
P++ T PV C D + TA CDK K +DL S ++ I QH+
Sbjct 79 ----PKAQQMHTGPVQDVCWSDDGTKVFTASCDKTAKMWDLNSNQSIQ--IAQHD 127
> cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor
Length=333
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 49/99 (49%), Gaps = 21/99 (21%)
Query 56 LACASWDRTCRVWQI---------------TTQPAAFGTNGQVLYKGNPQSLFTETAPVL 100
LA +SWD++ VW++ P F + +++ G + F +APVL
Sbjct 6 LAASSWDKSVTVWEVQHMGGNSVNTRFGKFLISPKVFN-DLLIVFIG---ASFQHSAPVL 61
Query 101 SCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
C ++ L + GCD ++K +D+ S ++ Q IG+H+
Sbjct 62 DCAISSDSRYLFSGGCDNELKMHDMSSRQS--QTIGRHD 98
> cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member
(npp-17); K14298 mRNA export factor
Length=373
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 58/124 (46%), Gaps = 19/124 (15%)
Query 20 STTESANYYSKQNSTELPNG-PSDSVSQLAWS---RDGQALACASWDRTCRVWQITTQPA 75
+TT +AN + QN L +G P D++ + +S +D LAC SWD T RVW
Sbjct 22 TTTPAAN--TTQNDDFLVDGAPEDTIQVIKFSPTPQDKPMLACGSWDGTIRVWMF----- 74
Query 76 AFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVI 135
N ++G Q AP+L + + + A DK+ + +DL S + V+
Sbjct 75 ----NDANTFEGKAQQ--NIPAPILDIAWIEDSSKIFIACADKEARLWDLASNQVA--VV 126
Query 136 GQHE 139
G H+
Sbjct 127 GTHD 130
> ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=339
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 48/105 (45%), Gaps = 17/105 (16%)
Query 35 ELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFT 94
EL N PSD +S L +S + L +SWD++ R++ NG ++ + F
Sbjct 10 ELSNPPSDGISNLRFSNNSDHLLVSSWDKSVRLYD---------ANGDLM-----RGEFK 55
Query 95 ETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
VL CC D + + D +V+ D +G+ V+G HE
Sbjct 56 HGGAVLDCCFHDDSSG-FSVCADTKVRRIDFNAGKE--DVLGTHE 97
> ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=340
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 47/105 (44%), Gaps = 17/105 (16%)
Query 35 ELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFT 94
EL N PSD +S L +S + L +SWD+ R++ ++T G+ L+ G
Sbjct 11 ELSNPPSDGISNLRFSNNSDHLLVSSWDKRVRLYDVSTN----SLKGEFLHGG------- 59
Query 95 ETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
VL CC D + G D +V+ G+ ++G H+
Sbjct 60 ---AVLDCCFHDDFSG-FSVGADYKVRRIVFNVGKE--DILGTHD 98
> cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3);
K02180 cell cycle arrest protein BUB3
Length=343
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 32/121 (26%), Positives = 57/121 (47%), Gaps = 16/121 (13%)
Query 20 STTESANYYSKQNSTELPNGPSDSVSQLAWSRDG--QALACASWDRTCRVWQITTQPAAF 77
ST ++A + N +P P +S++ + R+ + LA + WD TCRV+++
Sbjct 2 STYQAATIVAAPNEFRVPFPPFVQISKVQFQREAGSRLLAASGWDGTCRVYEV------- 54
Query 78 GTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
G G + + + +FT P+L+C ++ G D VK D+ +G G +G
Sbjct 55 GKLGDI----SEKLVFTHGKPLLTCTFAGYNKVAF-GGVDHNVKLADIETG--NGTQLGS 107
Query 138 H 138
H
Sbjct 108 H 108
> dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibited
by benzimidazoles 3 homolog (yeast); K02180 cell cycle
arrest protein BUB3
Length=326
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 50/112 (44%), Gaps = 24/112 (21%)
Query 32 NSTELPNGPSDSVSQLAWS-RDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQ 90
N +L GP DSVS + +S Q L +SWD + R LY +
Sbjct 5 NEFKLAQGPEDSVSAVKFSPSSSQFLLVSSWDGSVR-----------------LYDASAN 47
Query 91 SL---FTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
S+ + APVL C D T + G D Q+K +DL + + T ++G H+
Sbjct 48 SMRMKYQHLAPVLDCAFSDPTHA-WSGGLDSQLKTHDLNTDQDT--IVGTHD 96
> mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar protein
homolog B (Chlamydomonas)
Length=476
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 30/91 (32%), Positives = 39/91 (42%), Gaps = 13/91 (14%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
G D V+ L +S G LA AS DRT R+W + KG TAP
Sbjct 58 GHKDVVTSLQFSPQGNLLASASRDRTVRLWVLDR-------------KGKSSEFKAHTAP 104
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
V S Q+L+TA DK +K + + R
Sbjct 105 VRSVDFSADGQLLVTASEDKSIKVWSMFRQR 135
> ath:AT5G56130 transducin family protein / WD-40 repeat family
protein; K12880 THO complex subunit 3
Length=315
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
G V +AW+ +G LA S D+T R+W I +P + KG+ T++
Sbjct 18 GHKKKVHSVAWNSNGTKLASGSVDQTARIWNI--EPHGHSKAKDLELKGH-----TDSVD 70
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQV 134
L C + ++ TA DK V+ +D SG+ T QV
Sbjct 71 QL-CWDPKHSDLVATASGDKSVRLWDARSGKCTQQV 105
> sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of
the nuclear pore complex; required for polyadenylated RNA export
but not for protein import; homologous to S. pombe Rae1p;
K14298 mRNA export factor
Length=365
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 59/137 (43%), Gaps = 28/137 (20%)
Query 13 MAFAFGRSTTESA--------NYYSKQNSTELPNGPSDSVSQLAWS-RDGQALACASWDR 63
M+F F RS T SA N N + + DS+S +A+S + + +SWD
Sbjct 1 MSF-FNRSNTTSALGTSTAMANEKDLANDIVINSPAEDSISDIAFSPQQDFMFSASSWDG 59
Query 64 TCRVWQITTQPAAFGTNGQVLYKGNPQ--SLFTETAPVLSCCGGDTTQMLLTAGCDKQVK 121
R+W + G PQ + ++PVL + + + GCD +K
Sbjct 60 KVRIWDVQN--------------GVPQGRAQHESSSPVLCTRWSNDGTKVASGGCDNALK 105
Query 122 AYDLISGRTTGQVIGQH 138
YD+ SG+T Q IG H
Sbjct 106 LYDIASGQT--QQIGMH 120
> mmu:384605 Wdr88, Gm1429, Pqwd; WD repeat domain 88
Length=614
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 31/126 (24%), Positives = 53/126 (42%), Gaps = 11/126 (8%)
Query 5 CQTTESQKMAFAFGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRT 64
C +SQK+A + + S+ +P +++S ++ G L +SWD+
Sbjct 384 CFDPDSQKVASVSMDRCIKIWDITSRTTLFTIPKAHYNAISDCCFTSTGHFLCTSSWDKN 443
Query 65 CRVWQITTQPAAFGTNGQ--VLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKA 122
++W + T F G L KG+ V SCC + L++ G DK V
Sbjct 444 IKIWNVHT--GEFRNRGACVTLMKGH-------EGCVSSCCIARDSSFLISGGFDKTVAI 494
Query 123 YDLISG 128
+D+ G
Sbjct 495 WDVGGG 500
Score = 36.2 bits (82), Expect = 0.036, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 39/86 (45%), Gaps = 12/86 (13%)
Query 43 SVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVLSC 102
S++ + D Q +A S DR ++W IT++ F P++ + + C
Sbjct 379 SITSCCFDPDSQKVASVSMDRCIKIWDITSRTTLFTI---------PKAHYNAIS---DC 426
Query 103 CGGDTTQMLLTAGCDKQVKAYDLISG 128
C T L T+ DK +K +++ +G
Sbjct 427 CFTSTGHFLCTSSWDKNIKIWNVHTG 452
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 26/122 (21%), Positives = 49/122 (40%), Gaps = 31/122 (25%)
Query 40 PSDSVSQLAWSRDGQALACASWDRTCRVWQITTQP-------------AAFGTNGQV--- 83
P V + + + D + + +S+D+T R W + T F NG+
Sbjct 291 PKAPVLECSITADNRRIVASSYDKTVRAWDVETGQLLWKVRHDTFVVCCKFSPNGKFVLS 350
Query 84 -------LYKGNPQSLFT-------ETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+Y +P+++ T + SCC +Q + + D+ +K +D I+ R
Sbjct 351 ALDVDRGIYLMDPENITTVIHMKDHHQRSITSCCFDPDSQKVASVSMDRCIKIWD-ITSR 409
Query 130 TT 131
TT
Sbjct 410 TT 411
> cel:C14B1.4 tag-125; Temporarily Assigned Gene name family member
(tag-125); K14963 COMPASS component SWD3
Length=376
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 46/103 (44%), Gaps = 21/103 (20%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQ----VLYKGNPQSLFTET 96
SD VS ++++RDG +A S+D R+W NGQ ++ NP F +
Sbjct 213 SDPVSAVSFNRDGSLIASGSYDGLVRIWDT--------ANGQCIKTLVDDENPPVAFVKF 264
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
+P + +L + D +K +D G+T Q G HE
Sbjct 265 SP--------NGKYILASNLDSTLKLWDFSKGKTLKQYTG-HE 298
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 42/86 (48%), Gaps = 15/86 (17%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVLSCC 103
V+ +AWS D + + AS D+T ++++I T + KG+ +F CC
Sbjct 132 VNDIAWSSDSRCVVSASDDKTLKIFEIVT------SRMTKTLKGHNNYVF--------CC 177
Query 104 GGDTTQMLLTAGC-DKQVKAYDLISG 128
+ L+ +G D+ V+ +D+ +G
Sbjct 178 NFNPQSSLVVSGSFDESVRIWDVKTG 203
> cel:ZC302.2 hypothetical protein
Length=501
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 12/84 (14%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVL 100
SD ++ ++++ DG +A +S+D RVW AA G+ + L T+ APV
Sbjct 339 SDPITSISYNHDGNTMATSSYDGCIRVWD-----AASGSCLKTLVD-------TDHAPVT 386
Query 101 SCCGGDTTQMLLTAGCDKQVKAYD 124
C + LL+A D +K +D
Sbjct 387 FVCFSPNGKYLLSAQLDSSLKLWD 410
> ath:AT4G01860 transducin family protein / WD-40 repeat family
protein
Length=1308
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 33/112 (29%), Positives = 53/112 (47%), Gaps = 17/112 (15%)
Query 27 YYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYK 86
+YS + L G S+ ++ WS DG + S DR+ R+W+I +Q G V
Sbjct 222 HYSASHMLRL-TGHEGSIFRIVWSLDGSKIVSVSDDRSARIWEIDSQEVV----GPV--- 273
Query 87 GNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQH 138
LF + V CC D+ +++TAG D + + + G T +VI +H
Sbjct 274 -----LFGHSVRVWDCCISDS--LIVTAGEDCTCRVWG-VDG-TQLEVIKEH 316
> ath:AT1G69400 transducin family protein / WD-40 repeat family
protein; K02180 cell cycle arrest protein BUB3
Length=314
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 47/105 (44%), Gaps = 17/105 (16%)
Query 35 ELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFT 94
E N D+VS+L +S L ASWD R++ + + + N Q
Sbjct 7 EFENPIEDAVSRLRFSPQSNNLLVASWDSYLRLYDVESSSLSLELNSQ------------ 54
Query 95 ETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
A +L CC + + T+G D ++ YDL +G T IG+H+
Sbjct 55 --AALLDCCFENESTS-FTSGSDGFIRRYDLNAG--TVDTIGRHD 94
> ath:AT5G50120 transducin family protein / WD-40 repeat family
protein
Length=388
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 42 DSVSQLAWSRDGQALACASWDRTCRVWQIT 71
D+VS LA SRDG L SWDRT ++W+ T
Sbjct 166 DAVSGLALSRDGTLLYSVSWDRTLKIWRTT 195
> tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative
; K02180 cell cycle arrest protein BUB3
Length=332
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 47/108 (43%), Gaps = 18/108 (16%)
Query 33 STELPNGPSDSVSQLAW--SRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQ 90
S +L + P DS+S L + S LA SWD+T R++ + + L+K
Sbjct 2 SIDLRHEPRDSISSLCYAPSHGKSILAATSWDKTLRIYDVDAN--------EQLHK---- 49
Query 91 SLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQH 138
F P+L C + ++ G DKQV DL + + +G H
Sbjct 50 --FEFDMPLLDACFLGDSAKVVIGGLDKQVSLCDLQTEKVVS--LGSH 93
> mmu:69736 Nup37, 2410003L22Rik, 2810039M17Rik; nucleoporin 37;
K14302 nuclear pore complex protein Nup37
Length=326
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 17/99 (17%)
Query 31 QNSTELPNGPSDSVSQLAWS-RDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNP 89
+N ++ G SD ++ L + ++GQ LA S D TCR+W + +G
Sbjct 114 KNEYKVLEGHSDFINDLVFHPKEGQELASVSDDHTCRIWNL---------------EGKQ 158
Query 90 QSLFTETAPVLSCC-GGDTTQMLLTAGCDKQVKAYDLIS 127
+ F +P +S C + T L+ A + ++ YDL++
Sbjct 159 TAHFLLHSPGMSVCWHPEETFKLMVAEKNGTIRFYDLMA 197
> sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ribosomal
component essential for proper endonucleolytic cleavage
of the 35 S rRNA precursor at A0, A1, and A2 sites; contains
eight WD-repeats; PWP2 deletion leads to defects in cell
cycle and bud morphogenesis; K14558 periodic tryptophan
protein 2
Length=923
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 34/118 (28%), Positives = 51/118 (43%), Gaps = 17/118 (14%)
Query 15 FAFGRSTTESANYYSKQNSTEL--PNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITT 72
AFG S Y Q+ + + G DS + LA+S DG + AS D +VW IT+
Sbjct 317 LAFGSSKLGQLLVYEWQSESYILKQQGHFDSTNSLAYSPDGSRVVTASEDGKIKVWDITS 376
Query 73 QPAAFGTNGQVLYKGNPQSLFTE-TAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
G + F E T+ V + Q++ ++ D V+A+DLI R
Sbjct 377 --------------GFCLATFEEHTSSVTAVQFAKRGQVMFSSSLDGTVRAWDLIRYR 420
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 6/46 (13%)
Query 38 NGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQV 83
+G VS L++S++ LA ASWD+T R+W I FG + QV
Sbjct 470 SGHEGPVSCLSFSQENSVLASASWDKTIRIWSI------FGRSQQV 509
> xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat domain
5; K14963 COMPASS component SWD3
Length=334
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 44/89 (49%), Gaps = 19/89 (21%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
+S +AWS D L AS D+T ++W I++ G+ L KG+ +F
Sbjct 90 ISDVAWSSDSNLLVSASDDKTLKIWDISS--------GKCLKTLKGHSNYVF-------- 133
Query 102 CCGGDTTQMLLTAGC-DKQVKAYDLISGR 129
CC + L+ +G D+ V+ +D+ +G+
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGK 162
Score = 35.8 bits (81), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 20/93 (21%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY----KGNPQSLFTET 96
SD VS + ++RDG + +S+D CR+W + GQ L NP F +
Sbjct 171 SDPVSAVHFNRDGSLIVSSSYDGLCRIWDTAS--------GQCLKTLIDDDNPPVSFVKF 222
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+P + +L A D +K +D G+
Sbjct 223 SP--------NGKYILAATLDNTLKLWDYSKGK 247
> bbo:BBOV_III011180 17.m07962; WD domain, G-beta repeat containing
protein; K14556 U3 small nucleolar RNA-associated protein
12
Length=1005
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 33/121 (27%), Positives = 55/121 (45%), Gaps = 16/121 (13%)
Query 5 CQTTESQKMAFAFGRSTTESANYYSKQNSTELP-NGPSDSVSQLAWSRDGQALACASWDR 63
C +++ + +A A ST ++ YY+ L G V+ + S DG LA +S D+
Sbjct 638 CYSSDGRLLAVALEDSTIQT--YYADTLKPFLSLYGHKLPVTSIDISSDGALLASSSLDK 695
Query 64 TCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAY 123
T ++W + FG +SL +A V+ C + T L+T G D +K +
Sbjct 696 TTKIWGLD-----FGN--------IRRSLLGHSAAVVKCRWINGTHYLVTTGLDALIKMW 742
Query 124 D 124
D
Sbjct 743 D 743
> hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=436
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 40/91 (43%), Gaps = 13/91 (14%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
G D V+ + +S G LA AS DRT R+W I + F +K + TAP
Sbjct 16 GHKDVVTSVQFSPHGNLLASASRDRTVRLW-IPDKRGKFSE-----FKAH-------TAP 62
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
V S Q L TA DK +K + + R
Sbjct 63 VRSVDFSADGQFLATASEDKSIKVWSMYRQR 93
> sce:YBR175W SWD3, CPS30, SAF35; Essential subunit of the COMPASS
(Set1C) complex, which methylates histone H3 on lysine
4 and is required in transcriptional silencing near telomeres;
WD40 beta propeller superfamily member and ortholog of mammalian
WDR5; K14963 COMPASS component SWD3
Length=315
Score = 38.1 bits (87), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 40/95 (42%), Gaps = 14/95 (14%)
Query 45 SQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVLSCCG 104
S+L WS DGQ +A AS D + + ++ G + TAPV+S
Sbjct 59 SELCWSPDGQCIATASDDFSVEIIHLS--------------YGLLHTFIGHTAPVISLTF 104
Query 105 GDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQHE 139
+L T+ D+ +K +D ++G + E
Sbjct 105 NRKGNLLFTSSMDESIKIWDTLNGSLMKTISAHSE 139
> mmu:634555 nucleoporin Nup37-like
Length=326
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 47/99 (47%), Gaps = 17/99 (17%)
Query 31 QNSTELPNGPSDSVSQLAWS-RDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNP 89
+N ++ G SD ++ L + ++GQ LA S D TC++W + +G
Sbjct 114 KNEYKVLEGHSDFINDLVFHPKEGQELASVSDDHTCKIWNL---------------EGKQ 158
Query 90 QSLFTETAPVLSCC-GGDTTQMLLTAGCDKQVKAYDLIS 127
+ F +P +S C + T L+ A + ++ YDL++
Sbjct 159 TAHFLLHSPGMSVCWHPEETFKLMVAEKNGTIRFYDLMA 197
> xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 44/89 (49%), Gaps = 19/89 (21%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
+S +AWS D L AS D+T ++W +++ G+ L KG+ +F
Sbjct 90 ISDVAWSSDSNLLVSASDDKTLKIWDVSS--------GKCLKTLKGHSNYVF-------- 133
Query 102 CCGGDTTQMLLTAGC-DKQVKAYDLISGR 129
CC + L+ +G D+ V+ +D+ +G+
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGK 162
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 20/93 (21%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY----KGNPQSLFTET 96
SD VS + ++RDG + +S+D CR+W + GQ L NP F +
Sbjct 171 SDPVSAVHFNRDGSLIVSSSYDGLCRIWDTAS--------GQCLKTLIDDDNPPVSFVKF 222
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+P + +L A D +K +D G+
Sbjct 223 SP--------NGKYILAATLDNTLKLWDYSKGK 247
> mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3;
WD repeat domain 5; K14963 COMPASS component SWD3
Length=334
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 44/89 (49%), Gaps = 19/89 (21%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
+S +AWS D L AS D+T ++W +++ G+ L KG+ +F
Sbjct 90 ISDVAWSSDSNLLVSASDDKTLKIWDVSS--------GKCLKTLKGHSNYVF-------- 133
Query 102 CCGGDTTQMLLTAGC-DKQVKAYDLISGR 129
CC + L+ +G D+ V+ +D+ +G+
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGK 162
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 20/93 (21%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY----KGNPQSLFTET 96
SD VS + ++RDG + +S+D CR+W + GQ L NP F +
Sbjct 171 SDPVSAVHFNRDGSLIVSSSYDGLCRIWDTAS--------GQCLKTLIDDDNPPVSFVKF 222
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+P + +L A D +K +D G+
Sbjct 223 SP--------NGKYILAATLDNTLKLWDYSKGK 247
> hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPASS
component SWD3
Length=334
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 44/89 (49%), Gaps = 19/89 (21%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
+S +AWS D L AS D+T ++W +++ G+ L KG+ +F
Sbjct 90 ISDVAWSSDSNLLVSASDDKTLKIWDVSS--------GKCLKTLKGHSNYVF-------- 133
Query 102 CCGGDTTQMLLTAGC-DKQVKAYDLISGR 129
CC + L+ +G D+ V+ +D+ +G+
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGK 162
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 20/93 (21%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY----KGNPQSLFTET 96
SD VS + ++RDG + +S+D CR+W + GQ L NP F +
Sbjct 171 SDPVSAVHFNRDGSLIVSSSYDGLCRIWDTAS--------GQCLKTLIDDDNPPVSFVKF 222
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+P + +L A D +K +D G+
Sbjct 223 SP--------NGKYILAATLDNTLKLWDYSKGK 247
> dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 44/89 (49%), Gaps = 19/89 (21%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
+S +AWS D L AS D+T ++W +++ G+ L KG+ +F
Sbjct 90 ISDVAWSSDSNLLVSASDDKTLKIWDVSS--------GKCLKTLKGHSNYVF-------- 133
Query 102 CCGGDTTQMLLTAGC-DKQVKAYDLISGR 129
CC + L+ +G D+ V+ +D+ +G+
Sbjct 134 CCNFNPQSNLIVSGSFDESVRIWDVKTGK 162
Score = 35.8 bits (81), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 40/93 (43%), Gaps = 20/93 (21%)
Query 41 SDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY----KGNPQSLFTET 96
SD VS + ++RDG + +S+D CR+W + GQ L NP F +
Sbjct 171 SDPVSAVHFNRDGSLIVSSSYDGLCRIWDTAS--------GQCLKTLIDDDNPPVSFVKF 222
Query 97 APVLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+P + +L A D +K +D G+
Sbjct 223 SP--------NGKYILAATLDNTLKLWDYSKGK 247
> ath:AT1G47610 transducin family protein / WD-40 repeat family
protein
Length=351
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/53 (39%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Query 19 RSTTESANYYS-KQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQI 70
+S+ + +NY ++ T L SD+VS L+ + D L ASWDRT +VW+I
Sbjct 112 KSSVKPSNYVEVRRCRTALWIKHSDAVSCLSLAEDQGLLYSASWDRTVKVWRI 164
> tgo:TGME49_110310 WD domain, G-beta repeat-containing protein
Length=744
Score = 37.0 bits (84), Expect = 0.019, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 39/91 (42%), Gaps = 3/91 (3%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
GP+ V+ LA+ G A+ S D+T VW + T G+ S+F AP
Sbjct 296 GPTADVTALAFHPRGYAVLAVSADQTAWVWLLPTSTEKRGSRAPAPVT---LSVFVAGAP 352
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGR 129
+ +C G + + D V ++ +GR
Sbjct 353 LTACAFGAEGKTCIVGAADGSVNVFEAKTGR 383
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 42 DSVSQLAWSRDGQALACASWDRTCRVWQI 70
D+VS A+S DG+ LAC +D R++ I
Sbjct 207 DTVSATAFSLDGRLLACGCFDGDVRIYSI 235
> xla:379514 snrnp40-a, MGC64565, prp8bp, snrnp40, spf38, wdr57;
small nuclear ribonucleoprotein 40kDa (U5)
Length=337
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 47/129 (36%), Gaps = 29/129 (22%)
Query 26 NYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY 85
N Y ++ G S +V +L ++ DG L AS DRT +W T G +
Sbjct 74 NVYGDCDNYATLKGHSGAVMELHYNTDGSLLFSASTDRTVAIWDCETGERVKRLKGHTSF 133
Query 86 -------KGNPQSL----------------------FTETAPVLSCCGGDTTQMLLTAGC 116
+ PQ + F T VLS DT+ +++ G
Sbjct 134 VNSCYPARRGPQLICTGSDDGTVKLWDFRKKAAVQTFQNTYQVLSVTFNDTSDQIISGGI 193
Query 117 DKQVKAYDL 125
D +K +DL
Sbjct 194 DNDIKVWDL 202
Score = 31.6 bits (70), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 5/99 (5%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
G DSV+ L+ S +G L + D T RVW + +P A +++GN + E
Sbjct 213 GHGDSVTGLSLSSEGSYLLSNAMDNTVRVWDV--RPFAPKERCVKIFQGNVHNF--EKNL 268
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
+ S D ++ + D+ V +D S R ++ G
Sbjct 269 LRSSWSADGSK-IAAGSADRFVYVWDTTSRRVLYKLPGH 306
> xla:380404 snrnp40-b, MGC132119, MGC53216, prp8bp, spf38, wdr57;
small nuclear ribonucleoprotein 40kDa (U5); K12857 Prp8
binding protein
Length=337
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 47/129 (36%), Gaps = 29/129 (22%)
Query 26 NYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLY 85
N Y ++ G S +V +L ++ DG L AS D+T +W T G Y
Sbjct 74 NVYGDCDNYATLKGHSGAVMELHYNTDGSLLFSASTDKTVAIWDCQTGERVKRLKGHTSY 133
Query 86 -------KGNPQSL----------------------FTETAPVLSCCGGDTTQMLLTAGC 116
+ PQ + F T VLS DT+ +++ G
Sbjct 134 VNSCYPARRGPQLICTGSDDGTVKLWDFRKKAAVQTFQNTYQVLSVTFNDTSDQIISGGI 193
Query 117 DKQVKAYDL 125
D +K +DL
Sbjct 194 DNDIKVWDL 202
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 41/99 (41%), Gaps = 5/99 (5%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAP 98
G DSV+ L+ S +G L + D T RVW + +P A +++GN +
Sbjct 213 GHGDSVTGLSLSSEGSYLLSNAMDNTVRVWDV--RPFAPKERCVKIFQGNVHNF---EKN 267
Query 99 VLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
+L C + D+ V +D S R ++ G
Sbjct 268 LLRCSWSADGSKIAAGSADRFVYVWDTTSRRILYKLPGH 306
> ath:AT1G24130 transducin family protein / WD-40 repeat family
protein
Length=415
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 15/28 (53%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 42 DSVSQLAWSRDGQALACASWDRTCRVWQ 69
D+VS LA S+DG L ASWDR+ ++W+
Sbjct 193 DAVSSLALSQDGSLLYSASWDRSFKIWR 220
> tgo:TGME49_043540 U4/U6 small nuclear ribonucleoprotein, putative
(EC:2.6.1.45); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=711
Score = 36.6 bits (83), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 44/113 (38%), Gaps = 17/113 (15%)
Query 29 SKQNSTELP----NGPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL 84
S QNS EL G D V+++A+ G+ LA S D T R+W + Q G
Sbjct 444 SNQNSRELLVCKLEGHEDRVNRVAFHPSGRFLASTSHDETWRLWDVEKQQELLLQEGH-- 501
Query 85 YKGNPQSLFTETAPVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
A V ++ T V+ +DL +GRT ++G
Sbjct 502 -----------AAAVYGVSIHPDGSLIATTDLSGVVRVWDLRTGRTVMPLVGH 543
> bbo:BBOV_II003080 18.m06257; WD-repeat protein; K14855 ribosome
assembly protein 4
Length=548
Score = 36.2 bits (82), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 46/96 (47%), Gaps = 17/96 (17%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVL--YKGNPQSLFTETAPVLS 101
++ +A+S DG+ A AS+DRT R+W G G+ L +G+ ++ S
Sbjct 396 INHVAFSADGRLFASASFDRTVRIW--------CGITGRYLRTLRGHIGRVY---RIAWS 444
Query 102 CCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
CCG +L++ D +K +D +G+ + G
Sbjct 445 CCG----SLLISCSSDTTLKLWDAETGKLKFDLPGH 476
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITT 72
G S+SV + +S DG+ LA S D + R+W + T
Sbjct 126 GHSESVLCMDFSADGKLLATGSGDSSVRIWDLQT 159
> hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA
box binding protein (TBP)-associated factor, 100kDa; K03130
transcription initiation factor TFIID subunit 5
Length=800
Score = 35.8 bits (81), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVW 68
G +D+V L +SRDG+ LA S D T R+W
Sbjct 709 GHTDTVCSLRFSRDGEILASGSMDNTVRLW 738
> hsa:79023 NUP37, FLJ22618, MGC5585, p37; nucleoporin 37kDa;
K14302 nuclear pore complex protein Nup37
Length=326
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 47/99 (47%), Gaps = 17/99 (17%)
Query 31 QNSTELPNGPSDSVSQLAWS-RDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNP 89
+N ++ G +D ++ L + ++GQ +A S D TCR+W + +G
Sbjct 114 KNEYKVLEGHTDFINGLVFDPKEGQEIASVSDDHTCRIWNL---------------EGVQ 158
Query 90 QSLFTETAPVLSCC-GGDTTQMLLTAGCDKQVKAYDLIS 127
+ F +P +S C + T L+ A + ++ YDL++
Sbjct 159 TAHFVLHSPGMSVCWHPEETFKLMVAEKNGTIRFYDLLA 197
> mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase
II, TATA box binding protein (TBP)-associated factor; K03130
transcription initiation factor TFIID subunit 5
Length=801
Score = 35.8 bits (81), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVW 68
G +D+V L +SRDG+ LA S D T R+W
Sbjct 710 GHTDTVCSLRFSRDGEILASGSMDNTVRLW 739
> mmu:102162 Taf5l, 1110005N04Rik, AI849020; TAF5-like RNA polymerase
II, p300/CBP-associated factor (PCAF)-associated factor;
K03130 transcription initiation factor TFIID subunit 5
Length=589
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 37/83 (44%), Gaps = 15/83 (18%)
Query 56 LACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFT-ETAPVLSCCGGDTTQMLLTA 114
LA S D+T R+W +GN LFT PVLS + L +A
Sbjct 441 LATGSTDKTVRLWSAQ--------------QGNSVRLFTGHRGPVLSLSFSPNGKYLASA 486
Query 115 GCDKQVKAYDLISGRTTGQVIGQ 137
G D+++K +DL SG ++ G
Sbjct 487 GEDQRLKLWDLASGTLFKELRGH 509
Score = 34.7 bits (78), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITT---QPAAFGTNGQVL 84
G +DS++ LA+S D +A AS D + RVW I + A G++G+++
Sbjct 508 GHTDSITSLAFSPDSGLIASASMDNSVRVWDIRSTCCNTPADGSSGELV 556
> dre:553606 nle1, MGC110281, fc12b09, fc45f01, wu:fc12b09, wu:fc45f01,
zgc:110281; notchless homolog 1 (Drosophila); K14855
ribosome assembly protein 4
Length=476
Score = 35.4 bits (80), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 21/94 (22%), Positives = 44/94 (46%), Gaps = 13/94 (13%)
Query 44 VSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFTETAPVLSCC 103
V+++ +S D + +A AS+D++ ++W G G+ L SL PV
Sbjct 366 VNEVLFSPDTRLIASASFDKSIKIWD--------GKTGKYL-----NSLRGHVGPVYQVA 412
Query 104 GGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
+++L++ D +K +D+ +G+ + G
Sbjct 413 WSADSRLLVSGSSDSTLKVWDIKTGKLNADLPGH 446
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 7 TTESQKMAFAFGRSTTESANYYSKQNSTELPNGPSDSVSQLAWSRDGQALACASWDRTCR 66
+ +S+ + ST + + + + + +LP G +D V + WS DGQ +A D+ R
Sbjct 414 SADSRLLVSGSSDSTLKVWDIKTGKLNADLP-GHADEVFAVDWSPDGQRVASGGKDKCLR 472
Query 67 VWQ 69
+W+
Sbjct 473 IWR 475
> xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/CBP-associated
factor (PCAF)-associated factor, 65kDa; K03130
transcription initiation factor TFIID subunit 5
Length=587
Score = 35.4 bits (80), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 45/100 (45%), Gaps = 15/100 (15%)
Query 39 GPSDSVSQLAWSRDGQALACASWDRTCRVWQITTQPAAFGTNGQVLYKGNPQSLFT-ETA 97
G V + + + LA S D+T R+W +TQ +GN LFT
Sbjct 422 GHLSDVDCIKFHPNSNYLATGSSDKTVRLW--STQ------------QGNSVRLFTGHRG 467
Query 98 PVLSCCGGDTTQMLLTAGCDKQVKAYDLISGRTTGQVIGQ 137
PVL+ + L +AG D+++K +DL SG ++ G
Sbjct 468 PVLTLAFSPNGKYLASAGEDQRLKLWDLASGTQYKELRGH 507
Lambda K H
0.314 0.126 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40