bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0129_orf2
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
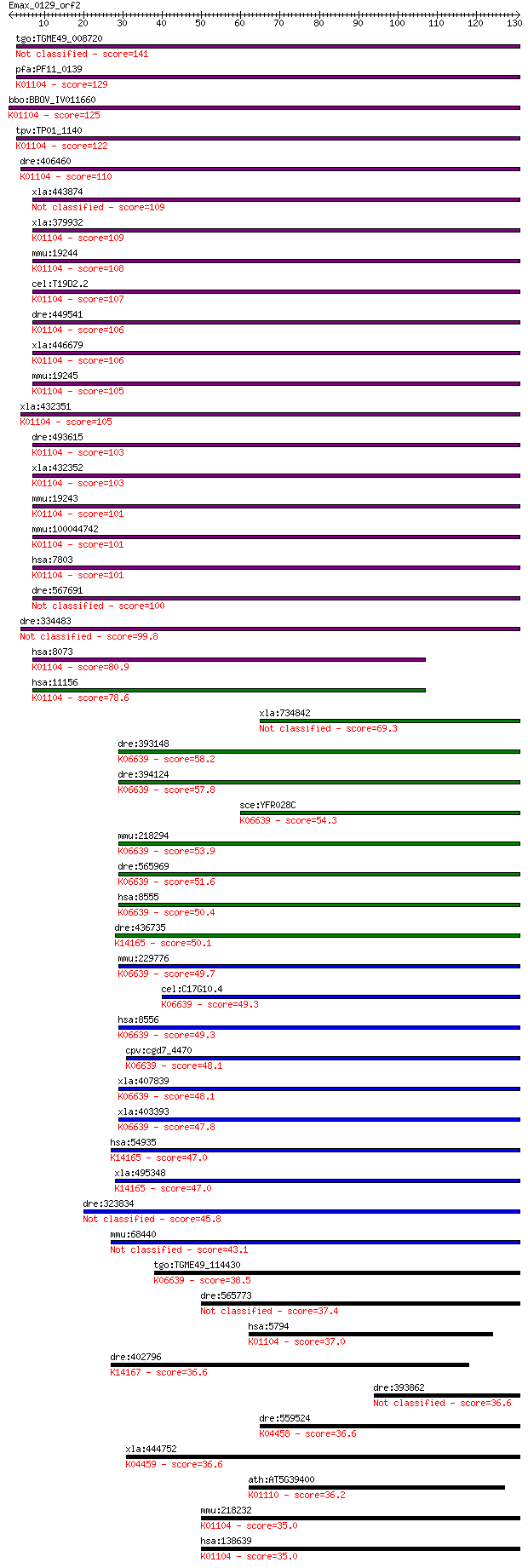
tgo:TGME49_008720 phosphatase, putative (EC:3.1.3.48) 141 5e-34
pfa:PF11_0139 PRL; protein tyrosine phosphatase; K01104 protei... 129 3e-30
bbo:BBOV_IV011660 23.m06279; tyrosine phosphatase (EC:3.1.3.48... 125 2e-29
tpv:TP01_1140 protein tyrosine phosphatase; K01104 protein-tyr... 122 2e-28
dre:406460 ptp4a3, wu:fc54b05, wu:fv52d11, zgc:77109; protein ... 110 9e-25
xla:443874 ptp4a2, MGC132077, MGC80084, hh13, hh7-2, ov-1, prl... 109 2e-24
xla:379932 ptp4a2, MGC53390; protein tyrosine phosphatase 4a2 ... 109 2e-24
mmu:19244 Ptp4a2, MGC102154, MGC103400, Prl-2; protein tyrosin... 108 5e-24
cel:T19D2.2 prl-1; hypothetical protein; K01104 protein-tyrosi... 107 1e-23
dre:449541 zgc:91861 (EC:3.1.3.48); K01104 protein-tyrosine ph... 106 2e-23
xla:446679 ptp4a1, MGC83351, hh72, prl-1, prl1, ptp(caax1), pt... 106 2e-23
mmu:19245 Ptp4a3, AV088979, Prl-3, pPtp4a3; protein tyrosine p... 105 4e-23
xla:432351 ptp4a3, prl-3, prl3, ptpcaax3; protein tyrosine pho... 105 4e-23
dre:493615 ptp4a1, zgc:101726; protein tyrosine phosphatase ty... 103 9e-23
xla:432352 prl-1; xPRL-1; K01104 protein-tyrosine phosphatase ... 103 1e-22
mmu:19243 Ptp4a1, AA415290, AU019864, C130021B01, MGC102117, M... 101 6e-22
mmu:100044742 protein tyrosine phosphatase type IVA 1-like; K0... 101 6e-22
hsa:7803 PTP4A1, DKFZp779M0721, HH72, PRL-1, PRL1, PTP(CAAX1),... 101 6e-22
dre:567691 fc14a08, wu:fc14a08; si:ch211-251p5.5 100 1e-21
dre:334483 PTP4A2, wu:fi84b06; zgc:101724 99.8 2e-21
hsa:8073 PTP4A2, HH13, HH7-2, HU-PP-1, OV-1, PRL-2, PRL2, PTP4... 80.9 9e-16
hsa:11156 PTP4A3, PRL-3, PRL-R, PRL3; protein tyrosine phospha... 78.6 4e-15
xla:734842 hypothetical protein MGC131305 69.3 3e-12
dre:393148 cdc14b, MGC55844, cdc14a, zgc:55844; CDC14 cell div... 58.2 6e-09
dre:394124 cdc14aa, CDC14A, MGC63654, zgc:63654; CDC14 cell di... 57.8 7e-09
sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell div... 54.3 1e-07
mmu:218294 Cdc14b, 2810432N10Rik, A530086E13Rik, AA472821, CDC... 53.9 1e-07
dre:565969 cdc14ab, si:dkey-168j9.1; CDC14 cell division cycle... 51.6 7e-07
hsa:8555 CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B; CDC14 cel... 50.4 1e-06
dre:436735 zgc:92902; K14165 dual specificity phosphatase [EC:... 50.1 2e-06
mmu:229776 Cdc14a, A830059A17Rik, CDC14A2, CDC14a1, Cdc14; CDC... 49.7 2e-06
cel:C17G10.4 cdc-14; Cell Division Cycle related family member... 49.3 3e-06
hsa:8556 CDC14A, cdc14, hCDC14; CDC14 cell division cycle 14 h... 49.3 3e-06
cpv:cgd7_4470 CDC14 phosphatase ; K06639 cell division cycle 1... 48.1 6e-06
xla:407839 cdc14b, MGC81657, cdc14beta, xcdc14b; CDC14 cell di... 48.1 7e-06
xla:403393 cdc14a, xcdc14a; CDC14 cell division cycle 14 homol... 47.8 9e-06
hsa:54935 DUSP23, DUSP25, FLJ20442, LDP-3, MOSP, RP11-190A12.1... 47.0 1e-05
xla:495348 dusp23; dual specificity phosphatase 23; K14165 dua... 47.0 1e-05
dre:323834 fc11c10, wu:fc11c10; si:dkeyp-95d10.1 45.8 3e-05
mmu:68440 Dusp23, 1300005N15Rik, LDP-3, MGC73633; dual specifi... 43.1 2e-04
tgo:TGME49_114430 dual specificity protein phosphatase CDC14A,... 38.5 0.005
dre:565773 ptpdc1, zgc:158271; protein tyrosine phosphatase do... 37.4 0.010
hsa:5794 PTPRH, FLJ39938, MGC133058, MGC133059, SAP1; protein ... 37.0 0.017
dre:402796 cdkn3; cyclin-dependent kinase inhibitor 3; K14167 ... 36.6 0.019
dre:393862 MGC77752; zgc:77752 36.6 0.020
dre:559524 ptpn5; protein tyrosine phosphatase, non-receptor t... 36.6 0.021
xla:444752 dusp5, MGC84792; dual specificity phosphatase 5; K0... 36.6 0.021
ath:AT5G39400 PTEN1; PTEN1; phosphatase; K01110 phosphatidylin... 36.2 0.027
mmu:218232 Ptpdc1, AI843923, AW456874, Naa-1; protein tyrosine... 35.0 0.057
hsa:138639 PTPDC1, FLJ42922, PTP9Q22; protein tyrosine phospha... 35.0 0.062
> tgo:TGME49_008720 phosphatase, putative (EC:3.1.3.48)
Length=483
Score = 141 bits (355), Expect = 5e-34, Method: Composition-based stats.
Identities = 67/128 (52%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 3 VLNVPTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIR 62
V+N PT IE+ K LI DAP+ +NL AY+ ++ VTDLVRTC TYD+ V+ +GIR
Sbjct 321 VMNTPTRIEAGRQKFLIFDAPSQENLPAYIEEMRAYEVTDLVRTCERTYDDKTVLASGIR 380
Query 63 VHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEA 122
H+L FPDGEAPP +VI W L + G +A+HCVAGLGR PVLVA++LI+ G +
Sbjct 381 PHELIFPDGEAPPDDVIDEWLTLCNAVSQQRGAIAIHCVAGLGRAPVLVAIALIEKGMDP 440
Query 123 EEAVNFIR 130
+A+ FIR
Sbjct 441 MDAIMFIR 448
> pfa:PF11_0139 PRL; protein tyrosine phosphatase; K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=218
Score = 129 bits (323), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 63/128 (49%), Positives = 84/128 (65%), Gaps = 0/128 (0%)
Query 3 VLNVPTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIR 62
VLN PT IE +K LILDAPTND L Y+ ++ VTDLVRTC TY++G + AGI
Sbjct 57 VLNHPTKIEHGKIKILILDAPTNDLLPLYIKEMKNYNVTDLVRTCERTYNDGEIQDAGIN 116
Query 63 VHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEA 122
VH+L FPDG+AP ++++ W + +AVHCVAGLGR PVL ++ LI+ G +
Sbjct 117 VHELIFPDGDAPTEDIVSNWLNIVNNVIKNNCAVAVHCVAGLGRAPVLASIVLIEFGMDP 176
Query 123 EEAVNFIR 130
+A+ FIR
Sbjct 177 IDAIVFIR 184
> bbo:BBOV_IV011660 23.m06279; tyrosine phosphatase (EC:3.1.3.48);
K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=172
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 58/130 (44%), Positives = 81/130 (62%), Gaps = 0/130 (0%)
Query 1 GMVLNVPTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAG 60
VLN PT IE ++ LILDAP N N++ YL +++ GVT LVRTC YD+ ++
Sbjct 9 SYVLNKPTKIEFHKLRILILDAPNNSNVKLYLHEMLDFGVTYLVRTCETNYDDSAIIEEN 68
Query 61 IRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGF 120
I + +L F DGE P E++A W L + A G +AVHCVAGLGR PVL ++L++ G
Sbjct 69 IAIKELIFNDGEPPSDEIVAEWLKLVKEVVASNGSVAVHCVAGLGRAPVLACIALVEYGM 128
Query 121 EAEEAVNFIR 130
+A+ F+R
Sbjct 129 HPLDAICFVR 138
> tpv:TP01_1140 protein tyrosine phosphatase; K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=168
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/128 (43%), Positives = 82/128 (64%), Gaps = 0/128 (0%)
Query 3 VLNVPTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIR 62
VLN PT IE + +K LILDAP N NL+ Y+ ++++ GV+ LVRTC Y++ ++ I
Sbjct 7 VLNKPTRIEYQKLKILILDAPNNSNLKLYIKEMLEFGVSCLVRTCESNYNDQLLLDNQIE 66
Query 63 VHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEA 122
V DL F DG+ PP +++ RW L +AVHCVAGLGR PVL ++L++ G +
Sbjct 67 VKDLFFNDGDPPPYDIVTRWLELIHHCLETNSAIAVHCVAGLGRAPVLACIALVEYGMQP 126
Query 123 EEAVNFIR 130
+A+ F+R
Sbjct 127 LDAICFVR 134
> dre:406460 ptp4a3, wu:fc54b05, wu:fv52d11, zgc:77109; protein
tyrosine phosphatase type IVA, member 3 (EC:3.1.3.48); K01104
protein-tyrosine phosphatase [EC:3.1.3.48]
Length=173
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 59/131 (45%), Positives = 82/131 (62%), Gaps = 4/131 (3%)
Query 4 LNVPTLIE--SRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGI 61
+N P +E + ++ LI PTN L +++ L + GVT +VR C TYD+ P+ GI
Sbjct 4 MNRPAPVEVCYKNMRFLITHNPTNSTLSSFIEDLKKYGVTTVVRVCEITYDKTPLEKNGI 63
Query 62 RVHDLTFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSG 119
V D F DG PP++V+ W +L + E G +AVHCVAGLGR PVLVAV+LI+SG
Sbjct 64 TVVDWPFDDGAPPPSKVVEDWLSLLKRRFIEEPGCCVAVHCVAGLGRAPVLVAVALIESG 123
Query 120 FEAEEAVNFIR 130
+ E+A+ FIR
Sbjct 124 MKYEDAIQFIR 134
> xla:443874 ptp4a2, MGC132077, MGC80084, hh13, hh7-2, ov-1, prl-2,
prl2, ptp-iv1b, ptpcaax2; protein tyrosine phosphatase
type IVA, member 2 (EC:3.1.3.48)
Length=167
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 76/126 (60%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI+V D
Sbjct 6 PVEISHECMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIQVLDW 65
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAEGG-VLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP +++ W L + + E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 66 PFDDGAPPPTQIVDDWLNLLKTKFREEAGCCIAVHCVAGLGRAPVLVALALIECGMKYED 125
Query 125 AVNFIR 130
AV FIR
Sbjct 126 AVQFIR 131
> xla:379932 ptp4a2, MGC53390; protein tyrosine phosphatase 4a2
(EC:3.1.3.48); K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=167
Score = 109 bits (272), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 76/126 (60%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI+V D
Sbjct 6 PVEISHECMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIQVLDW 65
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAEGG-VLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP +++ W L + + E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 66 PFDDGAPPPTQIVDDWLNLLKTKFREEAGCCIAVHCVAGLGRAPVLVALALIECGMKYED 125
Query 125 AVNFIR 130
AV FIR
Sbjct 126 AVQFIR 131
> mmu:19244 Ptp4a2, MGC102154, MGC103400, Prl-2; protein tyrosine
phosphatase 4a2 (EC:3.1.3.48); K01104 protein-tyrosine phosphatase
[EC:3.1.3.48]
Length=167
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI V D
Sbjct 6 PVEISYENMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIHVLDW 65
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP +++ W L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 66 PFDDGAPPPNQIVDDWLNLLKTKFREEPGCCVAVHCVAGLGRAPVLVALALIECGMKYED 125
Query 125 AVNFIR 130
AV FIR
Sbjct 126 AVQFIR 131
> cel:T19D2.2 prl-1; hypothetical protein; K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=190
Score = 107 bits (267), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 76/126 (60%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P+ I ++ LI D P N ++Q+Y+ +L + G +VR C PTYD + AGI V D
Sbjct 24 PSEIAWGKMRFLITDRPNNSSIQSYIEELEKHGARAVVRVCEPTYDTLALKEAGIDVLDW 83
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP EVI W L + E +AVHCVAGLGR PVLVA++LI++G + E+
Sbjct 84 QFSDGSPPPPEVIKSWFQLCMTSFKEHPDKSIAVHCVAGLGRAPVLVAIALIEAGMKYED 143
Query 125 AVNFIR 130
AV IR
Sbjct 144 AVEMIR 149
> dre:449541 zgc:91861 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=168
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I ++ LI PTN L + +L GV LVR C TYD+ PV GI V D
Sbjct 6 PVEITYECMRFLITHNPTNSQLAKFTEELKSFGVQTLVRVCESTYDKAPVEKEGIEVLDW 65
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAE-GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG +PP +++ W L + K E G +AVHCVAGLGR PVLVA++LI+ G E+
Sbjct 66 PFDDGCSPPEQIVDDWLNLLKCKFKDEPGCCIAVHCVAGLGRAPVLVAIALIECGMMYED 125
Query 125 AVNFIR 130
AV FIR
Sbjct 126 AVQFIR 131
> xla:446679 ptp4a1, MGC83351, hh72, prl-1, prl1, ptp(caax1),
ptpcaax1; protein tyrosine phosphatase type IVA, member 1 (EC:3.1.3.48);
K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=173
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 77/126 (61%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I + ++ LI PTN L ++ +L + GVT LVR C TYD V GI+V D
Sbjct 9 PVEITYKNMRFLITHNPTNATLNKFIEELKKYGVTTLVRVCEATYDTALVEKEGIQVLDW 68
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAE-GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P ++++ W L + + E G +AVHCVAGLGR PVLVA++LI+SG + E+
Sbjct 69 PFDDGAPPSSQIVDDWLNLLKVKFREEPGCCIAVHCVAGLGRAPVLVALALIESGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> mmu:19245 Ptp4a3, AV088979, Prl-3, pPtp4a3; protein tyrosine
phosphatase 4a3 (EC:3.1.3.48); K01104 protein-tyrosine phosphatase
[EC:3.1.3.48]
Length=173
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 55/126 (43%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + R ++ LI P+N L ++ L + G T +VR C TYD+ P+ GI V D
Sbjct 9 PVEVSYRHMRFLITHNPSNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEKDGITVVDW 68
Query 67 TFPDGEAPPAEVIARWRAL--AAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP +V+ W +L A G +AVHCVAGLGR PVLVA++LI+SG + E+
Sbjct 69 PFDDGAPPPGKVVEDWLSLLKAKFYNDPGSCVAVHCVAGLGRAPVLVALALIESGMKYED 128
Query 125 AVNFIR 130
A+ FIR
Sbjct 129 AIQFIR 134
> xla:432351 ptp4a3, prl-3, prl3, ptpcaax3; protein tyrosine phosphatase
type IVA, member 3 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=173
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 78/131 (59%), Gaps = 4/131 (3%)
Query 4 LNVPTLIE--SRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGI 61
+N P +E + ++ LI PTN L ++ L + G T +VR C TYD+ P+ GI
Sbjct 4 INRPAPVEVCYKNMRFLITHNPTNATLNTFIEDLKKYGATTVVRVCEITYDKTPLEKDGI 63
Query 62 RVHDLTFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSG 119
V D F DG PP++++ W L E G +AVHCVAGLGR PVLVA++LI+SG
Sbjct 64 TVMDWPFDDGAPPPSKIVDDWLNLLKTKFCEDPGCCVAVHCVAGLGRAPVLVALALIESG 123
Query 120 FEAEEAVNFIR 130
+ E+A+ FIR
Sbjct 124 MKYEDAIQFIR 134
> dre:493615 ptp4a1, zgc:101726; protein tyrosine phosphatase
type IVA, member 1 (EC:3.1.3.48); K01104 protein-tyrosine phosphatase
[EC:3.1.3.48]
Length=173
Score = 103 bits (258), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 56/126 (44%), Positives = 76/126 (60%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I + ++ LI PTN L ++ +L + GVT +VR C TYD VV GI+V D
Sbjct 9 PVEITYKNMRFLITHNPTNATLHKFIEELKKYGVTTVVRVCEATYDANLVVKEGIQVLDW 68
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAE-GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P +++ W L + + E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 69 PFDDGAPPSNQIVDDWLNLLRVKFREEPGCCIAVHCVAGLGRAPVLVALALIECGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> xla:432352 prl-1; xPRL-1; K01104 protein-tyrosine phosphatase
[EC:3.1.3.48]
Length=173
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/126 (44%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I + ++ LI PTN L ++ +L + GVT LVR C TYD V GI+V D
Sbjct 9 PVEITYKNMRFLITHNPTNATLNKFIEELKKFGVTTLVRVCEATYDTALVEKEGIQVLDW 68
Query 67 TFPDGEAPPAEVIARW-RALAAQAKAE-GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P +++ W L + + E G + VHCVAGLGR PVLVA++LI+SG + E+
Sbjct 69 PFDDGAPPSNQIVDDWLNLLKVKFREEPGCCITVHCVAGLGRAPVLVALALIESGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> mmu:19243 Ptp4a1, AA415290, AU019864, C130021B01, MGC102117,
MGC25304, Prl-1; protein tyrosine phosphatase 4a1 (EC:3.1.3.48);
K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=173
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 54/126 (42%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + + ++ LI PTN L ++ +L + GVT +VR C TYD V GI V D
Sbjct 9 PVEVTYKNMRFLITHNPTNATLNKFIEELKKYGVTTIVRVCEATYDTTLVEKEGIHVLDW 68
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P +++ W +L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 69 PFDDGAPPSNQIVDDWLSLVKIKFREEPGCCIAVHCVAGLGRAPVLVALALIEGGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> mmu:100044742 protein tyrosine phosphatase type IVA 1-like;
K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=173
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 54/126 (42%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + + ++ LI PTN L ++ +L + GVT +VR C TYD V GI V D
Sbjct 9 PVEVTYKNMRFLITHNPTNATLNKFIEELKKYGVTTIVRVCEATYDTTLVEKEGIHVLDW 68
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P +++ W +L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 69 PFDDGAPPSNQIVDDWLSLVKIKFREEPGCCIAVHCVAGLGRAPVLVALALIEGGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> hsa:7803 PTP4A1, DKFZp779M0721, HH72, PRL-1, PRL1, PTP(CAAX1),
PTPCAAX1; protein tyrosine phosphatase type IVA, member 1
(EC:3.1.3.48); K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=173
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 54/126 (42%), Positives = 73/126 (57%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + + ++ LI PTN L ++ +L + GVT +VR C TYD V GI V D
Sbjct 9 PVEVTYKNMRFLITHNPTNATLNKFIEELKKYGVTTIVRVCEATYDTTLVEKEGIHVLDW 68
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG P +++ W +L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 69 PFDDGAPPSNQIVDDWLSLVKIKFREEPGCCIAVHCVAGLGRAPVLVALALIEGGMKYED 128
Query 125 AVNFIR 130
AV FIR
Sbjct 129 AVQFIR 134
> dre:567691 fc14a08, wu:fc14a08; si:ch211-251p5.5
Length=173
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 51/126 (40%), Positives = 74/126 (58%), Gaps = 2/126 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + ++ +I PTN L ++ L + +VR C TYD+ P+ GI V D
Sbjct 9 PVEVCYNSMRFVITHNPTNQTLDTFIEDLKRYDAKTVVRVCESTYDKTPLEKHGITVMDW 68
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
F DG PP +++ W +L ++ +E G +AVHCVAGLGR PVLVAV+LI+ G + EE
Sbjct 69 PFDDGAPPPTKIVDDWISLLKKSFSEDPGCCVAVHCVAGLGRAPVLVAVALIEGGMKYEE 128
Query 125 AVNFIR 130
A++ IR
Sbjct 129 AIHLIR 134
> dre:334483 PTP4A2, wu:fi84b06; zgc:101724
Length=168
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 73/131 (55%), Gaps = 4/131 (3%)
Query 4 LNVPTLIE--SRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGI 61
+N P +E ++ LI PTN L + +L + V LVR C TYD V GI
Sbjct 1 MNRPAAVEISYDCMRFLITHNPTNSTLNKFTEELKKFEVNTLVRVCEATYDTALVQKEGI 60
Query 62 RVHDLTFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSG 119
+V D F DG +PP ++ W L E G +AVHCVAGLGR PVLVA++L++ G
Sbjct 61 QVFDWPFDDGASPPTRIVDDWLNLLKTKFREEPGCCIAVHCVAGLGRAPVLVALALLECG 120
Query 120 FEAEEAVNFIR 130
+ EEAV +IR
Sbjct 121 MKYEEAVMYIR 131
> hsa:8073 PTP4A2, HH13, HH7-2, HU-PP-1, OV-1, PRL-2, PRL2, PTP4A,
PTPCAAX2, ptp-IV1a, ptp-IV1b; protein tyrosine phosphatase
type IVA, member 2 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=142
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 44/102 (43%), Positives = 54/102 (52%), Gaps = 2/102 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI V D
Sbjct 6 PVEISYENMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIHVLDW 65
Query 67 TFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGR 106
F DG PP +++ W L E G +AVHCVAGLGR
Sbjct 66 PFDDGAPPPNQIVDDWLNLLKTKFREEPGCCVAVHCVAGLGR 107
> hsa:11156 PTP4A3, PRL-3, PRL-R, PRL3; protein tyrosine phosphatase
type IVA, member 3 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=148
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 42/102 (41%), Positives = 57/102 (55%), Gaps = 2/102 (1%)
Query 7 PTLIESRGVKCLILDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDL 66
P + + ++ LI PTN L ++ L + G T +VR C TYD+ P+ GI V D
Sbjct 9 PVEVSYKHMRFLITHNPTNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEKDGITVVDW 68
Query 67 TFPDGEAPPAEVIARWRAL--AAQAKAEGGVLAVHCVAGLGR 106
F DG PP +V+ W +L A +A G +AVHCVAGLGR
Sbjct 69 PFDDGAPPPGKVVEDWLSLVKAKFCEAPGSCVAVHCVAGLGR 110
> xla:734842 hypothetical protein MGC131305
Length=108
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 34/68 (50%), Positives = 45/68 (66%), Gaps = 2/68 (2%)
Query 65 DLTFPDGEAPPAEVIARWRALAAQAKAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEA 122
D F DG PP++++ W L E G +AVHCVAGLGR PVLVA++LI+SG +
Sbjct 2 DWPFDDGAPPPSKIVDDWLNLLKTKFCEDPGCCVAVHCVAGLGRAPVLVALALIESGMKY 61
Query 123 EEAVNFIR 130
E+A+ FIR
Sbjct 62 EDAIQFIR 69
> dre:393148 cdc14b, MGC55844, cdc14a, zgc:55844; CDC14 cell division
cycle 14 homolog B; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=404
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 35/103 (33%), Positives = 50/103 (48%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + VT ++R YD G + HDL F DG P +++R+ +
Sbjct 151 EAYFPYFRKHNVTTIIRLNKKMYDSKRFTDVGFKHHDLFFVDGSTPNDSIVSRFLHICEN 210
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
A GV+AVHC AGLGR L+ L+ A EA+ +IR
Sbjct 211 AD---GVIAVHCKAGLGRTGTLIGCYLMKHFRLTAAEAIAWIR 250
> dre:394124 cdc14aa, CDC14A, MGC63654, zgc:63654; CDC14 cell
division cycle 14 homolog A, a; K06639 cell division cycle 14
[EC:3.1.3.48]
Length=592
Score = 57.8 bits (138), Expect = 7e-09, Method: Composition-based stats.
Identities = 38/103 (36%), Positives = 50/103 (48%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + Q VTD+VR Y+ AG HDL F DG P + R+ +
Sbjct 208 EAYFSYFRQHNVTDVVRLNKKIYEGRRFTDAGFEHHDLFFVDGTTPSDLLTRRFLHICES 267
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
AK G +AVHC AGLGR L+ L+ F A EA+ + R
Sbjct 268 AK---GAVAVHCKAGLGRTGTLIGCYLMKHYRFTAPEAIAWTR 307
> sce:YFR028C CDC14, OAF3; Cdc14p (EC:3.1.3.48); K06639 cell division
cycle 14 [EC:3.1.3.48]
Length=551
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 60 GIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDS- 118
GI+ DL F DG P ++ + A GG +AVHC AGLGR L+ LI +
Sbjct 243 GIQHLDLIFEDGTCPDLSIVKNFVGAAETIIKRGGKIAVHCKAGLGRTGCLIGAHLIYTY 302
Query 119 GFEAEEAVNFIR 130
GF A E + F+R
Sbjct 303 GFTANECIGFLR 314
> mmu:218294 Cdc14b, 2810432N10Rik, A530086E13Rik, AA472821, CDC14B3,
Cdc14B1; CDC14 cell division cycle 14 homolog B (S.
cerevisiae) (EC:3.1.3.16 3.1.3.48); K06639 cell division cycle
14 [EC:3.1.3.48]
Length=448
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 47/103 (45%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+ Y+ VT ++R YD AG HDL FPDG P ++ + +
Sbjct 209 ETYIPYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFPDGSTPAESIVQEFLDICEN 268
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
K G +AVHC AGLGR L+ L+ A E++ ++R
Sbjct 269 VK---GAIAVHCKAGLGRTGTLIGCYLMKHYRMTAAESIAWLR 308
> dre:565969 cdc14ab, si:dkey-168j9.1; CDC14 cell division cycle
14 homolog A, b; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=510
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 37/103 (35%), Positives = 53/103 (51%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + VT +VR YD AG +DL F DG + P+++I R R L
Sbjct 226 EAYFPYFRKHNVTTIVRLNKKIYDAKRFTDAGFDHYDLFFVDG-STPSDIITR-RFLHI- 282
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
++ G +AVHC AGLGR L+ L+ F + EA+ +IR
Sbjct 283 CESTSGAVAVHCKAGLGRTGTLIGCYLMKHYRFTSAEAIAWIR 325
> hsa:8555 CDC14B, CDC14B3, Cdc14B1, Cdc14B2, hCDC14B; CDC14 cell
division cycle 14 homolog B (S. cerevisiae) (EC:3.1.3.16
3.1.3.48); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=461
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 47/103 (45%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+ Y+ VT ++R YD AG HDL F DG P ++ + +
Sbjct 209 ETYIQYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFADGSTPTDAIVKEFLDICEN 268
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
A+ G +AVHC AGLGR L+A ++ A E + ++R
Sbjct 269 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVR 308
> dre:436735 zgc:92902; K14165 dual specificity phosphatase [EC:3.1.3.16
3.1.3.48]
Length=152
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 51/107 (47%), Gaps = 8/107 (7%)
Query 28 LQAYLAQLVQVGVTDLV---RTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRA 84
+ A+ L+ G+ LV PP +D P + +H + D AP E I R+
Sbjct 25 MTAHYQYLLNSGIKHLVTLTERKPPDHDTCP----DLTLHHIKINDFCAPTFEQINRFLT 80
Query 85 LAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
+ +A A G +AVHC+ G GR ++A L+ S +A+N IR
Sbjct 81 IVEEANASGQAVAVHCLHGFGRTGTMLACYLVKSRKISGIDAINEIR 127
> mmu:229776 Cdc14a, A830059A17Rik, CDC14A2, CDC14a1, Cdc14; CDC14
cell division cycle 14 homolog A (S. cerevisiae) (EC:3.1.3.16
3.1.3.48); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=603
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + VT +VR Y+ AG +DL F DG P ++ R+ +
Sbjct 210 EAYFPYFKKNNVTTIVRLNKKIYEAKRFTDAGFEHYDLFFIDGSTPSDNIVRRFLNICEN 269
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
+ G +AVHC AGLGR L+A ++ F E + +IR
Sbjct 270 TE---GAIAVHCKAGLGRTGTLIACYVMKHYRFTHAEIIAWIR 309
> cel:C17G10.4 cdc-14; Cell Division Cycle related family member
(cdc-14); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=1063
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 33/92 (35%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query 40 VTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVH 99
V+ +VR YD AG DL F DG P E++ ++ + K GGV AVH
Sbjct 238 VSTIVRLNAKNYDASKFTKAGFDHVDLFFIDGSTPSDEIMLKFIKVVDNTK--GGV-AVH 294
Query 100 CVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIR 130
C AGLGR L+A ++ + G A E + ++R
Sbjct 295 CKAGLGRTGTLIACWMMKEYGLTAGECMGWLR 326
> hsa:8556 CDC14A, cdc14, hCDC14; CDC14 cell division cycle 14
homolog A (S. cerevisiae) (EC:3.1.3.16 3.1.3.48); K06639 cell
division cycle 14 [EC:3.1.3.48]
Length=594
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 48/103 (46%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + VT +VR Y+ AG +DL F DG P ++ R+ +
Sbjct 210 EAYFPYFKKHNVTAVVRLNKKIYEAKRFTDAGFEHYDLFFIDGSTPSDNIVRRFLNICEN 269
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
+ G +AVHC AGLGR L+A ++ F E + +IR
Sbjct 270 TE---GAIAVHCKAGLGRTGTLIACYVMKHYRFTHAEIIAWIR 309
> cpv:cgd7_4470 CDC14 phosphatase ; K06639 cell division cycle
14 [EC:3.1.3.48]
Length=453
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 49/101 (48%), Gaps = 4/101 (3%)
Query 31 YLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAK 90
Y+ ++ V+ ++R Y+ GI+ +L F DG PP ++ R+ L K
Sbjct 218 YIPIFKKLKVSTVIRLNKKQYESERFTNNGIKHEELFFIDGSCPPQNILNRFLELTENEK 277
Query 91 AEGGVLAVHCVAGLGR-GPVLVAVSLIDSGFEAEEAVNFIR 130
GV AVHC AGLGR G +L ++ + F A + + R
Sbjct 278 ---GVFAVHCKAGLGRTGTLLGCYAIKNYRFTASAWIGWNR 315
> xla:407839 cdc14b, MGC81657, cdc14beta, xcdc14b; CDC14 cell
division cycle 14 homolog B; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=452
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 46/103 (44%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + +T ++R YD A HDL F DG P ++ ++ +
Sbjct 209 EAYFPYFRKHHLTTIIRLNKKMYDANRFTDADFEHHDLFFVDGSTPSDAIVKKFLNICEN 268
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
A G +AVHC AGLGR L+ ++ A E + +IR
Sbjct 269 AD---GAIAVHCKAGLGRTGTLIGCYMMKHYRMTAAETIAWIR 308
> xla:403393 cdc14a, xcdc14a; CDC14 cell division cycle 14 homolog
A; K06639 cell division cycle 14 [EC:3.1.3.48]
Length=576
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 46/103 (44%), Gaps = 4/103 (3%)
Query 29 QAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 88
+AY + + ++R YD AG +DL F DG P ++ R+ L
Sbjct 208 EAYFPYFRKHNIRAVIRLNKKIYDAKRFTDAGFDHYDLFFVDGSTPSDGIVRRFLNLCEN 267
Query 89 AKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
G +AVHC AGLGR L+A ++ F E + +IR
Sbjct 268 TD---GAIAVHCKAGLGRTGTLIACYIMKHYRFTHSETIAWIR 307
> hsa:54935 DUSP23, DUSP25, FLJ20442, LDP-3, MOSP, RP11-190A12.1,
VHZ; dual specificity phosphatase 23 (EC:3.1.3.16 3.1.3.48);
K14165 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=150
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 51/108 (47%), Gaps = 8/108 (7%)
Query 27 NLQAYLAQLVQVGVTDLVRTC---PPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWR 83
L A+ L+ +GV LV PP D P G+ +H L PD P + I R+
Sbjct 23 RLPAHYQFLLDLGVRHLVSLTERGPPHSDSCP----GLTLHRLRIPDFCPPAPDQIDRFV 78
Query 84 ALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIR 130
+ +A A G + VHC G GR ++A L+ + G A +A+ IR
Sbjct 79 QIVDEANARGEAVGVHCALGFGRTGTMLACYLVKERGLAAGDAIAEIR 126
> xla:495348 dusp23; dual specificity phosphatase 23; K14165 dual
specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=151
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 50/107 (46%), Gaps = 8/107 (7%)
Query 28 LQAYLAQLVQVGVTDLV---RTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRA 84
L A+ L + G+ L+ PP +D P GI +H + D AP E I +
Sbjct 25 LPAHYEYLYENGIRHLITLTEHKPPYHDTCP----GITLHRIRIQDFCAPSLEQIKNFLK 80
Query 85 LAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLID-SGFEAEEAVNFIR 130
+ AK++G + VHC+ G GR ++A L+ +A+N IR
Sbjct 81 IVDDAKSKGEAVGVHCLHGFGRTGTMLACYLVKVRKITGVDAINEIR 127
> dre:323834 fc11c10, wu:fc11c10; si:dkeyp-95d10.1
Length=161
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 52/112 (46%), Gaps = 5/112 (4%)
Query 20 LDAPTNDNLQAYLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVI 79
L PT + YL + L+ PP Y++ P + +H ++ D P I
Sbjct 24 LARPTMVHHYRYLLDHGIKHLVSLLEIKPPNYEKCP----ELSLHQISIVDFTPPSRSQI 79
Query 80 ARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
++ ++ +A A+G +AVHC G GR ++A L+ S EEA+ IR
Sbjct 80 LQFLSIVEKANAKGEGVAVHCAHGHGRTGTMLACYLVKSRHLSGEEAIKEIR 131
> mmu:68440 Dusp23, 1300005N15Rik, LDP-3, MGC73633; dual specificity
phosphatase 23 (EC:3.1.3.16 3.1.3.48)
Length=150
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 49/108 (45%), Gaps = 8/108 (7%)
Query 27 NLQAYLAQLVQVGVTDLVRTC---PPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWR 83
L A+ L+ GV LV PP D P G+ +H + PD P E I ++
Sbjct 23 RLPAHYQFLLDQGVRHLVSLTERGPPHSDSCP----GLTLHRMRIPDFCPPSPEQIDQFV 78
Query 84 ALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIR 130
+ +A A G + VHC G GR ++A L+ + A +A+ IR
Sbjct 79 KIVDEANARGEAVGVHCALGFGRTGTMLACYLVKERALAAGDAIAEIR 126
> tgo:TGME49_114430 dual specificity protein phosphatase CDC14A,
putative (EC:3.1.3.16); K06639 cell division cycle 14 [EC:3.1.3.48]
Length=479
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 41/94 (43%), Gaps = 4/94 (4%)
Query 38 VGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLA 97
+G+ +VR YD I DL F DG P E+I +A + +A
Sbjct 226 LGIKTVVRLNKKQYDARKFTDRNIEHVDLFFVDGTCPSREII---QAFLQVVENRDHPIA 282
Query 98 VHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
VHC AGLGR L+ I + F A E + + R
Sbjct 283 VHCKAGLGRTGTLIGCYAIKNFKFPAVEWIGWNR 316
> dre:565773 ptpdc1, zgc:158271; protein tyrosine phosphatase
domain containing 1 (EC:3.1.3.-)
Length=713
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 41/82 (50%), Gaps = 2/82 (2%)
Query 50 TYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPV 109
TY + AGI ++ + D I + + A EG + AVHC AGLGR V
Sbjct 139 TYRPELFMEAGIYFYNFGWKDYGVASLTTILDMVKVMSFAMQEGKI-AVHCHAGLGRTGV 197
Query 110 LVAVSLI-DSGFEAEEAVNFIR 130
L+A L+ S A++A+ F+R
Sbjct 198 LIACFLVFTSRMSADQAILFVR 219
> hsa:5794 PTPRH, FLJ39938, MGC133058, MGC133059, SAP1; protein
tyrosine phosphatase, receptor type, H (EC:3.1.3.48); K01104
protein-tyrosine phosphatase [EC:3.1.3.48]
Length=937
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 62 RVHDLTFPDGEAP--PAEVIARWRALAA--QAKAEGGVLAVHCVAGLGRGPVLVAVSLID 117
+ H +PD P P ++A WR L EGG VHC AG+GR L+A+ ++
Sbjct 800 QFHYQAWPDHGVPSSPDTLLAFWRMLRQWLDQTMEGGPPIVHCSAGVGRTGTLIALDVLL 859
Query 118 SGFEAE 123
++E
Sbjct 860 RQLQSE 865
> dre:402796 cdkn3; cyclin-dependent kinase inhibitor 3; K14167
cyclin-dependent kinase inhibitor 3 [EC:3.1.3.16 3.1.3.48]
Length=208
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 45/104 (43%), Gaps = 18/104 (17%)
Query 27 NLQAYLAQLVQVGVTDLVRTCPP---TYDEGPVVLA-----GIRVHDLTFPDGEAPP--- 75
NLQ +A++ GV D+ C P +L G+RVH FPDG AP
Sbjct 54 NLQKDVAEMCDQGVEDVFVFCTRGELVRYRVPCLLEVYSQRGLRVHHFPFPDGGAPELYQ 113
Query 76 -AEVIARWR-ALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLID 117
+ V+ + L Q + +HC GLGR ++ A L+
Sbjct 114 CSCVLEELKDCLQNQRRT-----VIHCYGGLGRSGLIAACLLLQ 152
> dre:393862 MGC77752; zgc:77752
Length=464
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 26/38 (68%), Gaps = 1/38 (2%)
Query 94 GVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
G +AVHC AGLGR VL+A L+ + A EAV+++R
Sbjct 159 GKVAVHCHAGLGRTGVLIACYLVYTCRISASEAVHYVR 196
> dre:559524 ptpn5; protein tyrosine phosphatase, non-receptor
type 5; K04458 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=521
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 0/66 (0%)
Query 65 DLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 124
D PD P E++ QA G + VHC AG+GR +A S++ E
Sbjct 417 DQKTPDKAPPLLELVQEVEEARKQAPPNSGPVIVHCSAGIGRTGCFIATSILCQQLSNEG 476
Query 125 AVNFIR 130
V+ ++
Sbjct 477 VVDILK 482
> xla:444752 dusp5, MGC84792; dual specificity phosphatase 5;
K04459 dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=373
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 41/101 (40%), Gaps = 5/101 (4%)
Query 31 YLAQLVQVGVTDLVRTCPPTYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAK 90
+LA L + ++ R P + + I V D D + E I K
Sbjct 190 FLANLHITALLNVSRKSSPDFCKEQYSYKWIPVEDNHTADISSHFQEAID----FIDSVK 245
Query 91 AEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIR 130
GG + VHC AG+ R P + L+ + F EEA +I+
Sbjct 246 RAGGRVLVHCEAGISRSPTICMAYLMKTRKFHLEEAFEYIK 286
> ath:AT5G39400 PTEN1; PTEN1; phosphatase; K01110 phosphatidylinositol-3,4,5-trisphosphate
3-phosphatase [EC:3.1.3.67]
Length=412
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 32/72 (44%), Gaps = 12/72 (16%)
Query 62 RVHDLTFPDGEAPP-------AEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPVLVAVS 114
RV F D P E + W +L + + VHC+AG GR ++V+
Sbjct 112 RVERFPFDDNHVPSLKMIQLFCESVHSWLSLDPK-----NIAVVHCMAGKGRTGLMVSAY 166
Query 115 LIDSGFEAEEAV 126
L+ G AEEA+
Sbjct 167 LVYGGMSAEEAL 178
> mmu:218232 Ptpdc1, AI843923, AW456874, Naa-1; protein tyrosine
phosphatase domain containing 1 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=747
Score = 35.0 bits (79), Expect = 0.057, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 50 TYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPV 109
TY + AGI ++ + D I + A EG V AVHC AGLGR V
Sbjct 141 TYLPEAFMEAGIYFYNFGWKDYGVASLTAILDMVKVMTFALQEGKV-AVHCHAGLGRTGV 199
Query 110 LVAVSLI-DSGFEAEEAVNFIR 130
L+A L+ + A++A+ F+R
Sbjct 200 LIACYLVFATRMTADQAIIFVR 221
> hsa:138639 PTPDC1, FLJ42922, PTP9Q22; protein tyrosine phosphatase
domain containing 1 (EC:3.1.3.48); K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=806
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 50 TYDEGPVVLAGIRVHDLTFPDGEAPPAEVIARWRALAAQAKAEGGVLAVHCVAGLGRGPV 109
TY + AGI ++ + D I + A EG V A+HC AGLGR V
Sbjct 193 TYLPEAFMEAGIYFYNFGWKDYGVASLTTILDMVKVMTFALQEGKV-AIHCHAGLGRTGV 251
Query 110 LVAVSLI-DSGFEAEEAVNFIR 130
L+A L+ + A++A+ F+R
Sbjct 252 LIACYLVFATRMTADQAIIFVR 273
Lambda K H
0.321 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40