bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0263_orf1
Length=181
Score E
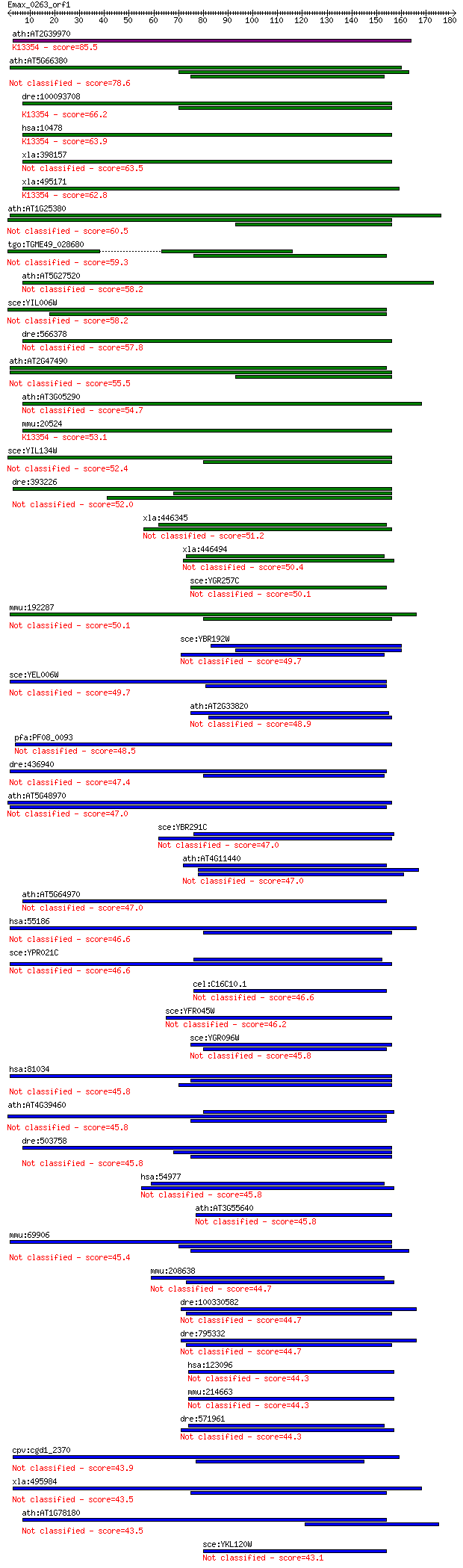
Sequences producing significant alignments: (Bits) Value
ath:AT2G39970 peroxisomal membrane protein (PMP36); K13354 sol... 85.5 1e-16
ath:AT5G66380 ATFOLT1 (ARABIDOPSIS THALIANA FOLATE TRANSPORTER... 78.6 1e-14
dre:100093708 zgc:162641; K13354 solute carrier family 25 (per... 66.2 6e-11
hsa:10478 SLC25A17, PMP34; solute carrier family 25 (mitochond... 63.9 3e-10
xla:398157 slc25a17-b, pmp34; solute carrier family 25 (mitoch... 63.5 4e-10
xla:495171 slc25a17-a, pmp34, slc25a17; solute carrier family ... 62.8 7e-10
ath:AT1G25380 mitochondrial substrate carrier family protein; ... 60.5 3e-09
tgo:TGME49_028680 peroxisomal membrane protein, putative 59.3 7e-09
ath:AT5G27520 PNC2; PNC2 (PEROXISOMAL ADENINE NUCLEOTIDE CARRI... 58.2 1e-08
sce:YIL006W YIA6, NDT1; Mitochondrial NAD+ transporter, involv... 58.2 2e-08
dre:566378 solute carrier family 25 (mitochondrial carrier; pe... 57.8 2e-08
ath:AT2G47490 mitochondrial substrate carrier family protein; ... 55.5 9e-08
ath:AT3G05290 PNC1; PNC1 (PEROXISOMAL ADENINE NUCLEOTIDE CARRI... 54.7 2e-07
mmu:20524 Slc25a17, 34kDa, 47kDa, PMP34, Pmp47; solute carrier... 53.1 5e-07
sce:YIL134W FLX1; Flx1p; K15115 solute carrier family 25 (mito... 52.4 9e-07
dre:393226 slc25a32a, MGC55610, fi40c12, mftc, wu:fi40c12, zgc... 52.0 1e-06
xla:446345 slc25a40, MGC81365; solute carrier family 25, membe... 51.2 2e-06
xla:446494 slc25a38, MGC80014; solute carrier family 25, membe... 50.4 3e-06
sce:YGR257C MTM1; Mitochondrial protein of the mitochondrial c... 50.1 4e-06
mmu:192287 Slc25a36, C330005L02Rik; solute carrier family 25, ... 50.1 4e-06
sce:YBR192W RIM2, PYT1; Mitochondrial pyrimidine nucleotide tr... 49.7 6e-06
sce:YEL006W YEA6, NDT2; Putative mitochondrial NAD+ transporte... 49.7 6e-06
ath:AT2G33820 MBAC1; MBAC1; L-histidine transmembrane transpor... 48.9 9e-06
pfa:PF08_0093 mitochondrial carrier protein, putative 48.5 1e-05
dre:436940 slc25a36a, zgc:92447; solute carrier family 25, mem... 47.4 2e-05
ath:AT5G48970 mitochondrial substrate carrier family protein; ... 47.0 3e-05
sce:YBR291C CTP1; Ctp1p; K15100 solute carrier family 25 (mito... 47.0 3e-05
ath:AT4G11440 binding 47.0 3e-05
ath:AT5G64970 mitochondrial substrate carrier family protein 47.0 3e-05
hsa:55186 SLC25A36, DKFZp564C053, FLJ10618; solute carrier fam... 46.6 4e-05
sce:YPR021C AGC1; Agc1p; K15105 solute carrier family 25 (mito... 46.6 4e-05
cel:C16C10.1 hypothetical protein; K15119 solute carrier famil... 46.6 5e-05
sce:YFR045W Putative mitochondrial transport protein; null mut... 46.2 6e-05
sce:YGR096W TPC1; Mitochondrial membrane transporter that medi... 45.8 7e-05
hsa:81034 SLC25A32, FLJ23872, MFT, MFTC; solute carrier family... 45.8 7e-05
ath:AT4G39460 SAMC1; SAMC1 (S-ADENOSYLMETHIONINE CARRIER 1); S... 45.8 8e-05
dre:503758 slc25a32b, zgc:110786; solute carrier family 25, me... 45.8 8e-05
hsa:54977 SLC25A38, FLJ20551, FLJ22703; solute carrier family ... 45.8 8e-05
ath:AT3G55640 mitochondrial substrate carrier family protein 45.8 8e-05
mmu:69906 Slc25a32, 2610043O12Rik, Mftc; solute carrier family... 45.4 1e-04
mmu:208638 Slc25a38, AV019094, BC010801, MGC18873; solute carr... 44.7 2e-04
dre:100330582 hypothetical protein LOC100330582; K15100 solute... 44.7 2e-04
dre:795332 similar to Tricarboxylate transport protein, mitoch... 44.7 2e-04
hsa:123096 SLC25A29, C14orf69, CACL, FLJ38975; solute carrier ... 44.3 2e-04
mmu:214663 Slc25a29, C030003J19Rik, CACL, mCACL; solute carrie... 44.3 2e-04
dre:571961 slc25a38b; Solute carrier family 25 member 38-B; K1... 44.3 3e-04
cpv:cgd1_2370 mitochondrial carrier protein, Flx1p like mitoch... 43.9 3e-04
xla:495984 slc25a32; solute carrier family 25, member 32; K151... 43.5 3e-04
ath:AT1G78180 binding 43.5 4e-04
sce:YKL120W OAC1; Mitochondrial inner membrane transporter, tr... 43.1 5e-04
> ath:AT2G39970 peroxisomal membrane protein (PMP36); K13354 solute
carrier family 25 (peroxisomal adenine nucleotide transporter),
member 17
Length=331
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 52/165 (31%), Positives = 90/165 (54%), Gaps = 31/165 (18%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G+ G++P L++V NP++QF LY+ + KL K+R + + S+N
Sbjct 188 GFWKGVIPTLIMVSNPSMQFMLYETMLTKLK--KKR----ALKGSNN------------- 228
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD----YSSSVRSL 118
VT E+FL+G AKL A + TYP LVVK+R Q+K D Y ++ ++
Sbjct 229 --------VTALETFLLGAVAKLGATVTTYPLLVVKSRLQAKQVTTGDKRQQYKGTLDAI 280
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLF 163
+ ++ EGL G + G+ +K+ ++L++A++F + E+L+ LL
Sbjct 281 LKMIRYEGLYGFYKGMSTKIVQSVLAAAVLFMIKEELVKGAKLLL 325
> ath:AT5G66380 ATFOLT1 (ARABIDOPSIS THALIANA FOLATE TRANSPORTER
1); binding / folic acid transporter; K15115 solute carrier
family 25 (mitochondrial folate transporter), member 32
Length=308
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 57/161 (35%), Positives = 85/161 (52%), Gaps = 26/161 (16%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
R GIVP LVLV + AIQFT Y+ LR ++ LK+R R + E + N L+S
Sbjct 163 RALYKGIVPGLVLVSHGAIQFTAYEELRKIIVDLKERRRKS--ESTDNLLNS-------- 212
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
A+ +G ++K+AA L+TYP+ V++ R Q + + + SL +I
Sbjct 213 ------------ADYAALGGSSKVAAVLLTYPFQVIRARLQQRP-STNGIPRYIDSLHVI 259
Query 122 LET---EGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLV 159
ET EGL G + GL + L + +S+I F VYE +L L+
Sbjct 260 RETARYEGLRGFYRGLTANLLKNVPASSITFIVYENVLKLL 300
Score = 37.7 bits (86), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 48/96 (50%), Gaps = 5/96 (5%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ--SKVYNVKDYSSSVRSLIMILETEGL 127
+++PA A L T P +VKTR Q + ++ + YS + + I++ EG
Sbjct 103 KLSPALHLASAAEAGALVCLCTNPIWLVKTRLQLQTPLHQTQPYSGLLDAFRTIVKEEGP 162
Query 128 SGLFAGLPSKLTATLLS-SAIMFTVYEQLLPLVVLL 162
L+ G+ L L+S AI FT YE+L ++V L
Sbjct 163 RALYKGIVPGLV--LVSHGAIQFTAYEELRKIIVDL 196
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 36/81 (44%), Gaps = 3/81 (3%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQ---SKVYNVKDYSSSVRSLIMILETEGLSGLF 131
E+ G A A + VV+TR Q + ++ Y ++ ++ I EGL GL+
Sbjct 8 ENATAGAVAGFATVAAMHSLDVVRTRFQVNDGRGSSLPTYKNTAHAVFTIARLEGLRGLY 67
Query 132 AGLPSKLTATLLSSAIMFTVY 152
AG + + +S + F Y
Sbjct 68 AGFFPAVIGSTVSWGLYFFFY 88
> dre:100093708 zgc:162641; K13354 solute carrier family 25 (peroxisomal
adenine nucleotide transporter), member 17
Length=312
Score = 66.2 bits (160), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 51/163 (31%), Positives = 79/163 (48%), Gaps = 51/163 (31%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LVLNPA+QF +Y+ L+ +++ R RE SS
Sbjct 170 GTFPSLLLVLNPAVQFMIYEGLKRQIL------RGVHRELSS------------------ 205
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT----------RAQSKVYNVKDYSSSVR 116
E FL+G AK A +TYP V++ QS++ N S+R
Sbjct 206 -------VEVFLIGAVAKAVATTITYPLQTVQSVLRFGQHGQPAGQSRLLN------SLR 252
Query 117 SLIMILETE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
S++ +L G+ GLF GL +KL T+L++A+MF +YE++
Sbjct 253 SVMYLLINRVRKWGILGLFKGLEAKLLQTVLTAALMFLLYEKI 295
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVKD-----YSSSVRSLIMIL 122
R T ++G+AA + LVT P VV TR Q + +D Y+ + + I+
Sbjct 100 RSTAGRDLIIGIAAGVVNVLVTTPLWVVNTRLKLQGAKFRNEDIQPTHYNGIKDAFVQIM 159
Query 123 ETEGLSGLFAG-LPSKLTATLLSSAIMFTVYEQL 155
EG+ L+ G PS L +L+ A+ F +YE L
Sbjct 160 RQEGVGALWNGTFPSLL--LVLNPAVQFMIYEGL 191
> hsa:10478 SLC25A17, PMP34; solute carrier family 25 (mitochondrial
carrier; peroxisomal membrane protein, 34kDa), member
17; K13354 solute carrier family 25 (peroxisomal adenine nucleotide
transporter), member 17
Length=307
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 78/156 (50%), Gaps = 39/156 (25%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LV NPAIQF Y+ L+ +L++
Sbjct 168 GTFPSLLLVFNPAIQFMFYEGLKRQLLKK------------------------------- 196
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT--RAQSKVYNVKDYS-SSVRSLIMILE 123
R++++ + F++G AK A VTYP V++ R N ++ + S+R+++ +L
Sbjct 197 -RMKLSSLDVFIIGAVAKAIATTVTYPLQTVQSILRFGRHRLNPENRTLGSLRNILYLLH 255
Query 124 TE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
G+ GL+ GL +KL T+L++A+MF VYE+L
Sbjct 256 QRVRRFGIMGLYKGLEAKLLQTVLTAALMFLVYEKL 291
> xla:398157 slc25a17-b, pmp34; solute carrier family 25 (mitochondrial
carrier; peroxisomal membrane protein, 34kDa), member
17
Length=310
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 74/156 (47%), Gaps = 39/156 (25%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LV NPAIQF Y+ L+ +L++ +
Sbjct 175 GTFPSLLLVFNPAIQFMFYEALKRQLLKGQTE---------------------------- 206
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT---RAQSKVYNVKDYSSSVRSLIMILE 123
+T E F++G AK A VTYP V++ Q K+ K S+ +I +L+
Sbjct 207 ----LTAMEVFVIGAIAKAIATAVTYPLQTVQSVLRFGQEKLNPEKRPLGSLHRVIYLLQ 262
Query 124 TE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
G+ GL+ GL +KL T+L++A+MF VYE+L
Sbjct 263 QRVKRWGIFGLYKGLEAKLLQTVLTAALMFLVYEKL 298
> xla:495171 slc25a17-a, pmp34, slc25a17; solute carrier family
25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa),
member 17; K13354 solute carrier family 25 (peroxisomal
adenine nucleotide transporter), member 17
Length=310
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 49/159 (30%), Positives = 75/159 (47%), Gaps = 39/159 (24%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LV NPAIQF Y+ L+ +L++ +
Sbjct 175 GTFPSLLLVFNPAIQFMFYEALKRQLLKGQTE---------------------------- 206
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT---RAQSKVYNVKDYSSSVRSLIMILE 123
+T E F++G AK A +TYP V++ Q K K S+R +I +L+
Sbjct 207 ----LTAMEVFVIGAIAKAIATALTYPLQTVQSVLRFGQEKRNPEKRPLGSLRRVIYLLQ 262
Query 124 TE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPL 158
G+ GL+ GL +KL T+L++A+MF VYE+L L
Sbjct 263 QRVKRWGILGLYKGLEAKLLQTVLTAALMFLVYEKLTSL 301
> ath:AT1G25380 mitochondrial substrate carrier family protein;
K15115 solute carrier family 25 (mitochondrial folate transporter),
member 32
Length=363
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 44/177 (24%), Positives = 83/177 (46%), Gaps = 35/177 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG +GI+P+L V + AIQF Y+ ++ + ++ ++ S ++ S G++
Sbjct 174 RGLYSGILPSLAGVSHVAIQFPAYEKIKQYMAKM-------------DNTSVENLSPGNV 220
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVK-DYSSSVRSL 118
+ AK+ A+++TYP+ V++ + Q ++ N + YS + +
Sbjct 221 A---------------IASSIAKVIASILTYPHEVIRAKLQEQGQIRNAETKYSGVIDCI 265
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLRVPTYRKRD 175
+ +EG+ GL+ G + L T S+ I FT YE +L FR + P + D
Sbjct 266 TKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEMML----RFFRQVVPPETNRSD 318
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 65/159 (40%), Gaps = 38/159 (23%)
Query 1 FRGWLAGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTG 59
+RG G+ P ++ +L N A+ F++Y L+ L SS G
Sbjct 76 YRGMYRGLSPTIIALLPNWAVYFSVYGKLKDVL----------------------QSSDG 113
Query 60 SLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVKDYSSSVRS 117
L + + + A A ++ T P VVKTR Q V Y S + +
Sbjct 114 KL----------SIGSNMIAAAGAGAATSIATNPLWVVKTRLMTQGIRPGVVPYKSVMSA 163
Query 118 LIMILETEGLSGLFAG-LPSKLTATLLSSAIMFTVYEQL 155
I EG+ GL++G LPS A + AI F YE++
Sbjct 164 FSRICHEEGVRGLYSGILPS--LAGVSHVAIQFPAYEKI 200
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 9/70 (12%)
Query 93 PYLVVKTRAQSKVYNVKDYSSS-------VRSLIMILETEGLSGLFAGLPSKLTATLLSS 145
P V+KTR Q V + + +S + SL I++ EG G++ GL + A L +
Sbjct 37 PLDVIKTRLQ--VLGLPEAPASGQRGGVIITSLKNIIKEEGYRGMYRGLSPTIIALLPNW 94
Query 146 AIMFTVYEQL 155
A+ F+VY +L
Sbjct 95 AVYFSVYGKL 104
> tgo:TGME49_028680 peroxisomal membrane protein, putative
Length=520
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 29/53 (54%), Positives = 37/53 (69%), Gaps = 0/53 (0%)
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV 115
+ R R ++ E+F +GL AKL A L TYPYLVVKTRAQ+K++NV D S V
Sbjct 463 TQRTPRGALSGGEAFAIGLIAKLCATLATYPYLVVKTRAQTKLHNVHDNSGIV 515
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/37 (59%), Positives = 28/37 (75%), Gaps = 0/37 (0%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQ 37
RGW AG++PAL+LV NPAIQF LYD L+ L +K+
Sbjct 342 LRGWFAGLLPALMLVSNPAIQFVLYDFLKDTLTAVKE 378
Score = 36.2 bits (82), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
S ++ +AA + + + + P+ V TR K+ + + R L IL EGL G FAGL
Sbjct 293 SIIIAVAAGICSTIASNPFWVANTRI--KLGASRHTTDVWRMLGYILRREGLRGWFAGL- 349
Query 136 SKLTATLLSS--AIMFTVYE 153
L A +L S AI F +Y+
Sbjct 350 --LPALMLVSNPAIQFVLYD 367
> ath:AT5G27520 PNC2; PNC2 (PEROXISOMAL ADENINE NUCLEOTIDE CARRIER
2); ADP transmembrane transporter/ ATP transmembrane transporter/
binding
Length=321
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 48/178 (26%), Positives = 86/178 (48%), Gaps = 34/178 (19%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G+ +L+L NPAIQ+T++D L+ L++ + + S + SS
Sbjct 162 GLGISLLLTSNPAIQYTVFDQLKQNLLE---KGKAKSNKDSSP----------------- 201
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ----SKVYNVKDYSSSVRSLI--- 119
+ ++ +F++G +K AA ++TYP + K Q SK K +R I
Sbjct 202 --VVLSAFMAFVLGAVSKSAATVITYPAIRCKVMIQAADDSKENEAKKPRKRIRKTIPGV 259
Query 120 --MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLP---LVVLLFRSLRVPTYR 172
I + EG+ G F GL +++ T+LSSA++ + E++ +++L R+L V R
Sbjct 260 VYAIWKKEGILGFFKGLQAQILKTVLSSALLLMIKEKITATTWILILAIRTLFVTKAR 317
> sce:YIL006W YIA6, NDT1; Mitochondrial NAD+ transporter, involved
in the transport of NAD+ into the mitochondria (see also
YEA6); member of the mitochondrial carrier subfamily; disputed
role as a pyruvate transporter; has putative mouse and
human orthologs; K15115 solute carrier family 25 (mitochondrial
folate transporter), member 32
Length=373
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 47/158 (29%), Positives = 76/158 (48%), Gaps = 36/158 (22%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
F+ AG+VP+L+ + + AI F +Y+ LK R+ SRE ++NS++ Q S
Sbjct 233 FKALYAGLVPSLLGLFHVAIHFPIYE-------DLKVRFHCYSRENNTNSINLQRLIMAS 285
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLI 119
VS K+ A+ VTYP+ +++TR Q K D S+ R L
Sbjct 286 SVS--------------------KMIASAVTYPHEILRTRMQLK----SDIPDSIQRRLF 321
Query 120 MILET----EGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+++ EGL G ++G + L T+ +SAI +E
Sbjct 322 PLIKATYAQEGLKGFYSGFTTNLVRTIPASAITLVSFE 359
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (41%), Gaps = 13/148 (8%)
Query 18 PAIQFTLYDHLRAKLMQLKQR-WRTASREPSSNSLSS---QHSSTGSLVSDRMLRIRVTP 73
P Q+ DH L+ +Q + S + SS + +SST S++ LR + P
Sbjct 15 PEQQYCSADHEEPLLLHEEQLIFPDHSSQLSSADIIEPIKMNSSTESIIGT-TLRKKWVP 73
Query 74 AESF----LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILET----E 125
S L G A + + P V KTR Q++ + + R ++ L T E
Sbjct 74 LSSTQITALSGAFAGFLSGVAVCPLDVAKTRLQAQGLQTRFENPYYRGIMGTLSTIVRDE 133
Query 126 GLSGLFAGLPSKLTATLLSSAIMFTVYE 153
G GL+ GL + + I F+VYE
Sbjct 134 GPRGLYKGLVPIVLGYFPTWMIYFSVYE 161
> dre:566378 solute carrier family 25 (mitochondrial carrier;
peroxisomal membrane protein, 34kDa), member 17-like
Length=303
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 47/156 (30%), Positives = 73/156 (46%), Gaps = 39/156 (25%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G +P+LVLV NPA+QF Y+ ++ R A R
Sbjct 164 GTLPSLVLVFNPAVQFMFYEAMK----------RKAGRGGR------------------- 194
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSS---SVRSLIMIL- 122
+++ E FL+G AK A TYP V+ + Y D S+R+++ +L
Sbjct 195 ---KISSFEIFLIGAIAKAIATTATYPLQTVQAILRFGQYKSDDKGGLVGSLRNVVSLLM 251
Query 123 ---ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+ GL GL+ GL +KL T+L++A+MF VYE++
Sbjct 252 DRIKRHGLLGLYKGLEAKLLQTVLTAALMFVVYEKI 287
> ath:AT2G47490 mitochondrial substrate carrier family protein;
K15115 solute carrier family 25 (mitochondrial folate transporter),
member 32
Length=312
Score = 55.5 bits (132), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 40/153 (26%), Positives = 72/153 (47%), Gaps = 29/153 (18%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG +G+VPAL + + AIQF Y+ ++ L + + S ++L+++ + S
Sbjct 170 RGLYSGLVPALAGISHVAIQFPTYEMIKVYLAKKGDK--------SVDNLNARDVAVASS 221
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI-M 120
+ AK+ A+ +TYP+ VV+ R Q + ++ + S VR I
Sbjct 222 I--------------------AKIFASTLTYPHEVVRARLQEQGHHSEKRYSGVRDCIKK 261
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+ E +G G + G + L T ++ I FT +E
Sbjct 262 VFEKDGFPGFYRGCATNLLRTTPAAVITFTSFE 294
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 67/157 (42%), Gaps = 36/157 (22%)
Query 2 RGWLAGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
RG G+ P ++ +L N AI FT+YD L++ L S+ H
Sbjct 73 RGLYRGLSPTVMALLSNWAIYFTMYDQLKSFL------------------CSNDH----- 109
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK--DYSSSVRSL 118
+++ + L A A + T P VVKTR Q++ V Y S+ +L
Sbjct 110 ---------KLSVGANVLAASGAGAATTIATNPLWVVKTRLQTQGMRVGIVPYKSTFSAL 160
Query 119 IMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
I EG+ GL++GL L A + AI F YE +
Sbjct 161 RRIAYEEGIRGLYSGLVPAL-AGISHVAIQFPTYEMI 196
Score = 37.0 bits (84), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 93 PYLVVKTRAQSKVY------NVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSA 146
P V+KTR Q N+K S V SL I + EG+ GL+ GL + A L + A
Sbjct 33 PLDVIKTRFQVHGLPKLGDANIKG-SLIVGSLEQIFKREGMRGLYRGLSPTVMALLSNWA 91
Query 147 IMFTVYEQL 155
I FT+Y+QL
Sbjct 92 IYFTMYDQL 100
> ath:AT3G05290 PNC1; PNC1 (PEROXISOMAL ADENINE NUCLEOTIDE CARRIER
1); ADP transmembrane transporter/ ATP transmembrane transporter/
binding
Length=322
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 47/170 (27%), Positives = 82/170 (48%), Gaps = 31/170 (18%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G+ +L+L NPAIQ+T++D L+ L LKQ+ + +++ S+ ++S M
Sbjct 160 GLGISLLLTSNPAIQYTVFDQLKQHL--LKQK-----------NAKAENGSSPVVLSAFM 206
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ----SKVYNVKDYSSSVRSLI--- 119
+F++G +K A ++TYP + K Q SK K R I
Sbjct 207 ---------AFVLGAVSKSVATVLTYPAIRCKVMIQAADESKENETKKPRRRTRKTIPGV 257
Query 120 --MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLR 167
I EG+ G F GL +++ T+LSSA++ + E++ +L ++R
Sbjct 258 VYAIWRKEGMLGFFKGLQAQILKTVLSSALLLMIKEKITATTWILILAIR 307
> mmu:20524 Slc25a17, 34kDa, 47kDa, PMP34, Pmp47; solute carrier
family 25 (mitochondrial carrier, peroxisomal membrane protein),
member 17; K13354 solute carrier family 25 (peroxisomal
adenine nucleotide transporter), member 17
Length=307
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 78/156 (50%), Gaps = 39/156 (25%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G P+L+LV NPAIQF Y+ L+ +L++
Sbjct 168 GTFPSLLLVFNPAIQFMFYEGLKRQLLKK------------------------------- 196
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT--RAQSKVYNVKDYS-SSVRSLIMILE 123
R++++ + F++G AK A VTYP V++ R N ++ + S+R+++ +L
Sbjct 197 -RMKLSSLDVFIIGAIAKAIATTVTYPMQTVQSILRFGRHRLNPENRTLGSLRNVLSLLH 255
Query 124 TE----GLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
G+ GL+ GL +KL T+L++A+MF VYE+L
Sbjct 256 QRVKRFGIMGLYKGLEAKLLQTVLTAALMFLVYEKL 291
> sce:YIL134W FLX1; Flx1p; K15115 solute carrier family 25 (mitochondrial
folate transporter), member 32
Length=311
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 68/156 (43%), Gaps = 29/156 (18%)
Query 1 FRGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
F+G G+VPAL V A+ F +YD LKQR RE
Sbjct 180 FQGLWKGLVPALFGVSQGALYFAVYD-------TLKQRKLRRKRENG------------- 219
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI- 119
L I +T E+ + K+ + + YP+ ++K+ QS + + + LI
Sbjct 220 ------LDIHLTNLETIEITSLGKMVSVTLVYPFQLLKSNLQS--FRANEQKFRLFPLIK 271
Query 120 MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+I+ +G GL+ GL + L + S+ I F VYE L
Sbjct 272 LIIANDGFVGLYKGLSANLVRAIPSTCITFCVYENL 307
Score = 36.6 bits (83), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 2/77 (2%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKD-YSSSVRSLIMILETEGLSGLFAGLPSKL 138
G ++ L A++T P V+KTR S + Y+S + +L T+G GL+ GL L
Sbjct 132 GASSGLMTAILTNPIWVIKTRIMSTSKGAQGAYTSMYNGVQQLLRTDGFQGLWKGLVPAL 191
Query 139 TATLLSSAIMFTVYEQL 155
+ A+ F VY+ L
Sbjct 192 FG-VSQGALYFAVYDTL 207
> dre:393226 slc25a32a, MGC55610, fi40c12, mftc, wu:fi40c12, zgc:55610;
solute carrier family 25, member 32a; K15115 solute
carrier family 25 (mitochondrial folate transporter), member
32
Length=324
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 63/153 (41%), Gaps = 30/153 (19%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G G VP LV + A+QF Y+ L+ + + K + PS + LS
Sbjct 193 GLYRGFVPGLVGTSHAALQFMTYEGLKREQNKCK-------KMPSESLLS---------- 235
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMIL 122
P E + +K+ A VTYPY VV+ R Q + N YS V +
Sbjct 236 ----------PLEYIAIAAISKIFAVAVTYPYQVVRARLQDQHNN---YSGIVDVMRRTW 282
Query 123 ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ G + G+ L + + I F V+E +
Sbjct 283 SNEGVEGFYKGMVPNLVRVIPACCITFLVFENV 315
Score = 32.7 bits (73), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 6/92 (6%)
Query 68 RIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV----KDYSSSVRSLIMILE 123
+ ++ E + A + +T P V KTR + YN K Y + +L+ I
Sbjct 129 QTELSACEHLVSAAEAGILTLCLTNPVWVTKTRLVLQ-YNADPSRKQYKGMMDALVKIYR 187
Query 124 TEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ GL+ G L T +A+ F YE L
Sbjct 188 HEGIPGLYRGFVPGLVGT-SHAALQFMTYEGL 218
Score = 28.9 bits (63), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 25/117 (21%), Positives = 52/117 (44%), Gaps = 2/117 (1%)
Query 41 TASREPSSNSLSSQHSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTR 100
T SR+ + + S+ S+ ++ + + E+ GLA + + +V +P ++K R
Sbjct 4 TISRQRHAAVAADSSGSSFSITANLLQLSKHIKYENLAAGLAGGVISTMVLHPLDLIKIR 63
Query 101 -AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
A S ++ Y + + I + EG+ GL+ G+ + S + F Y +
Sbjct 64 FAVSDGLKMRPQYDGMLDCMKTIWKLEGIRGLYQGVTPNIWGAGSSWGLYFLFYNAI 120
> xla:446345 slc25a40, MGC81365; solute carrier family 25, member
40; K15119 solute carrier family 25, member 39/40
Length=341
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 51/99 (51%), Gaps = 7/99 (7%)
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKV-------YNVKDYSSS 114
+ R ++ T A SF G + AA+VT P+ VVKTR Q +V Y+ K SS+
Sbjct 225 LCQRYNTLQPTFAISFTAGAVSGSIAAIVTLPFDVVKTRRQVEVGELEVFTYSHKRSSST 284
Query 115 VRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+ + I+ G GLFAGL +L + AIM + YE
Sbjct 285 WKLMSAIVAENGFGGLFAGLVPRLIKVAPACAIMISTYE 323
Score = 37.0 bits (84), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 23/107 (21%), Positives = 52/107 (48%), Gaps = 7/107 (6%)
Query 56 SSTGSLVSD------RMLRIRV-TPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV 108
SS G+L++ +++IR+ ++ F+ G L+ + + ++
Sbjct 23 SSVGALLTSFLVTPLDVVKIRLQAQSKPFIKGKCFVYCNGLMDHLCMCTNGNGKAWYKAP 82
Query 109 KDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+ ++ + + I+ +EG+ L++GLP L + ++ I FT Y+QL
Sbjct 83 GHFRGTMDAFVQIIRSEGIKSLWSGLPPTLVMAVPATVIYFTFYDQL 129
> xla:446494 slc25a38, MGC80014; solute carrier family 25, member
38; K15118 solute carrier family 25, member 38
Length=302
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 45/80 (56%), Gaps = 7/80 (8%)
Query 73 PAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFA 132
P ES ++G ++ AA+ P+ VVKTR +S Y Y+S +L I +TEG GLF+
Sbjct 121 PLESVMLGAGSRTVAAVCMLPFTVVKTRYESGKYG---YNSVYGALKAIYKTEGPRGLFS 177
Query 133 GLPSKLTATLLSSAIMFTVY 152
G LTATL+ A +Y
Sbjct 178 G----LTATLMRDAPFSGIY 193
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 51/86 (59%), Gaps = 5/86 (5%)
Query 72 TPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLF 131
+P +F G+ A + A++ T P V+KT Q + N K Y + + + I T+GL+G F
Sbjct 214 SPVLNFSCGIVAGILASVATQPADVIKTHMQ--LANEK-YHWTGKVALNIYRTQGLTGFF 270
Query 132 -AGLPSKLTATLLSSAIMFTVYEQLL 156
G+P L TL+ +A+ +TVYEQ++
Sbjct 271 QGGVPRALRRTLM-AAMAWTVYEQMM 295
> sce:YGR257C MTM1; Mitochondrial protein of the mitochondrial
carrier family, involved in activating mitochondrial Sod2p
probably by facilitating insertion of an essential manganese
cofactor; K15119 solute carrier family 25, member 39/40
Length=366
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 43/85 (50%), Gaps = 6/85 (7%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIM------ILETEGLS 128
SF G + + AA+ T+P+ V KTR Q + N D RS M I TEGL+
Sbjct 270 NSFASGCISGMIAAICTHPFDVGKTRWQISMMNNSDPKGGNRSRNMFKFLETIWRTEGLA 329
Query 129 GLFAGLPSKLTATLLSSAIMFTVYE 153
L+ GL +++ S AIM + YE
Sbjct 330 ALYTGLAARVIKIRPSCAIMISSYE 354
> mmu:192287 Slc25a36, C330005L02Rik; solute carrier family 25,
member 36; K15116 solute carrier family 25, member 33/36
Length=311
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 42/164 (25%), Positives = 71/164 (43%), Gaps = 26/164 (15%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG+ G+ + + I F +Y+ ++ KL++ K + E S S
Sbjct 174 RGFYRGMSASYAGISETVIHFVIYESIKQKLLECKTASMMETDEESVKEASD-------- 225
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+R+ + A S K A + YP+ VV+TR + + Y S ++L +I
Sbjct 226 ----FVRMMLAAATS-------KTCATTIAYPHEVVRTRLREEG---TKYRSFFQTLSLI 271
Query 122 LETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
++ EG L+ GL + L + ++AIM YE LVV L
Sbjct 272 VQEEGYGSLYRGLTTHLVRQIPNTAIMMATYE----LVVYLLNG 311
Score = 32.3 bits (72), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 35/95 (36%), Gaps = 21/95 (22%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVR-------------------SLIM 120
G A++T P VVKTR QS +V Y S V+ L
Sbjct 13 GGCGGTVGAILTCPLEVVKTRLQSS--SVTLYISEVQLNTMAGASVNRVVSPGPLHCLKA 70
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
ILE EG LF GL L S AI F Y
Sbjct 71 ILEKEGPRSLFRGLGPNLVGVAPSRAIYFAAYSNC 105
> sce:YBR192W RIM2, PYT1; Mitochondrial pyrimidine nucleotide
transporter; imports pyrimidine nucleoside triphosphates and
exports pyrimidine nucleoside monophosphates; member of the
mitochondrial carrier family; K15116 solute carrier family
25, member 33/36
Length=377
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 83 AKLAAALVTYPYLVVKTRAQS--KVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTA 140
AK A++ TYP+ VV+TR + K + Y+ V+S +I++ EGL +++GL L
Sbjct 298 AKFVASIATYPHEVVRTRLRQTPKENGKRKYTGLVQSFKVIIKEEGLFSMYSGLTPHLMR 357
Query 141 TLLSSAIMFTVYEQLLPLV 159
T+ +S IMF +E ++ L+
Sbjct 358 TVPNSIIMFGTWEIVIRLL 376
Score = 32.7 bits (73), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 93 PYLVVKTRAQ---SKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMF 149
P ++KTR Q + +V+ Y +S L ++ EG +GL+ GL + + + + +
Sbjct 195 PIWLIKTRVQLDKAGKTSVRQYKNSWDCLKSVIRNEGFTGLYKGLSASYLGS-VEGILQW 253
Query 150 TVYEQLLPLV 159
+YEQ+ L+
Sbjct 254 LLYEQMKRLI 263
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 45/108 (41%), Gaps = 26/108 (24%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVY---------NVKDYSSSVRSLIMI 121
V P F+ G +A A+VT P+ +VKTR QS ++ N+ S+ +S+ +
Sbjct 50 VKPWVHFVAGGIGGMAGAVVTCPFDLVKTRLQSDIFLKAYKSQAVNISKGSTRPKSINYV 109
Query 122 LET-----------------EGLSGLFAGLPSKLTATLLSSAIMFTVY 152
++ EG LF GL L + + +I F Y
Sbjct 110 IQAGTHFKETLGIIGNVYKQEGFRSLFKGLGPNLVGVIPARSINFFTY 157
> sce:YEL006W YEA6, NDT2; Putative mitochondrial NAD+ transporter,
member of the mitochondrial carrier subfamily (see also
YIA6); has putative human ortholog; K15115 solute carrier family
25 (mitochondrial folate transporter), member 32
Length=335
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 43/157 (27%), Positives = 78/157 (49%), Gaps = 33/157 (21%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
+ AG+VPAL+ +LN AIQF LY++L+ + + S + S++ SS L
Sbjct 196 KALYAGLVPALLGMLNVAIQFPLYENLKIRF------GYSESTDVSTDVTSSNFQ---KL 246
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLIM 120
+ ML +K+ A+ VTYP+ +++TR Q K D ++V R L+
Sbjct 247 ILASML---------------SKMVASTVTYPHEILRTRMQLK----SDLPNTVQRHLLP 287
Query 121 ILE----TEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
+++ EG +G ++G + L T+ ++ + +E
Sbjct 288 LIKITYRQEGFAGFYSGFATNLVRTVPAAVVTLVSFE 324
Score = 34.7 bits (78), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 39/78 (50%), Gaps = 6/78 (7%)
Query 81 LAAKLAAALVTYPYLVVKTRAQSK-----VYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
L+ L+A LV P+ V KTR Q++ + + Y + I + EG +GL+ GL
Sbjct 47 LSGALSAMLVC-PFDVAKTRLQAQGLQNMTHQSQHYKGFFGTFATIFKDEGAAGLYKGLQ 105
Query 136 SKLTATLLSSAIMFTVYE 153
+ + + I F+VY+
Sbjct 106 PTVLGYIPTLMIYFSVYD 123
> ath:AT2G33820 MBAC1; MBAC1; L-histidine transmembrane transporter/
L-lysine transmembrane transporter/ L-ornithine transmembrane
transporter/ arginine transmembrane transporter/ binding;
K15109 solute carrier family 25 (mitochondrial carnitine/acylcarnitine
transporter), member 20/29
Length=311
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKD--YSSSVRSLIMILETEGLSGLFA 132
+ ++ G+ A LA V +P+ VK + Q +V+ Y + + IL+TEG+ GL+
Sbjct 16 KEYVAGMMAGLATVAVGHPFDTVKVKLQKHNTDVQGLRYKNGLHCASRILQTEGVKGLYR 75
Query 133 GLPSKLTATLLSSAIMFTVYEQ 154
G S S++MF +Y Q
Sbjct 76 GATSSFMGMAFESSLMFGIYSQ 97
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 8/82 (9%)
Query 82 AAKLAAALVTY---PYLVVKTRAQ-----SKVYNVKDYSSSVRSLIMILETEGLSGLFAG 133
+A A++++ P +VK R Q S V N + Y+S + + ++ +G++G+F G
Sbjct 119 SAMFGGAIISFVLCPTELVKCRMQIQGTDSLVPNFRRYNSPLDCAVQTVKNDGVTGIFRG 178
Query 134 LPSKLTATLLSSAIMFTVYEQL 155
+ L +A+ FTVYE L
Sbjct 179 GSATLLRECTGNAVFFTVYEYL 200
> pfa:PF08_0093 mitochondrial carrier protein, putative
Length=540
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 38/152 (25%), Positives = 68/152 (44%), Gaps = 22/152 (14%)
Query 4 WLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVS 63
+ G + +L+L + A+QF Y++L S + +N L +++ S V
Sbjct 406 FYKGFLASLLLTPHVALQFYTYEYLT----------HFFSSQYINNILQNKNIHFSSDVL 455
Query 64 DRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILE 123
++ L FL G +K A + TYP +K R Q ++ N Y + L+ I +
Sbjct 456 NKFL--------PFLYGALSKYIAIIFTYPLYTIKMRQQVQMKNYGFY----KVLLNIFK 503
Query 124 TEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ + G+ L L + I+F ++E L
Sbjct 504 CEGIRSYYTGINMHLLRNCLQNGILFFIFEYL 535
> dre:436940 slc25a36a, zgc:92447; solute carrier family 25, member
36a; K15116 solute carrier family 25, member 33/36
Length=311
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 70/156 (44%), Gaps = 30/156 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG+ G+ + + I F +Y+ ++ KL++ K +++++ + S
Sbjct 174 RGFYRGMSASYAGISETVIHFVIYESIKRKLIEHK----------ANSNMDDEDES---- 219
Query 62 VSDRMLRIRVTPAESFL-MGLAA---KLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRS 117
V A F+ M LAA K A + YP+ V++TR + + Y S ++
Sbjct 220 ---------VKDASDFVGMMLAAATSKTCATSIAYPHEVIRTRLREEG---SKYRSFFQT 267
Query 118 LIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
L M+ EG L+ GL + L + ++AIM YE
Sbjct 268 LNMVFREEGYRALYRGLTTHLVRQIPNTAIMMCTYE 303
Score = 34.3 bits (77), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 36/92 (39%), Gaps = 21/92 (22%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVR-------------------SLIM 120
G A++T P VVKTR QS +V Y S V+ L +
Sbjct 13 GGCGGTVGAILTCPLEVVKTRLQSS--SVTFYISEVQLSTVNGASVARMAPPGPLHCLKL 70
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVY 152
ILE EG LF GL L S AI F Y
Sbjct 71 ILEKEGPRSLFRGLGPNLVGVAPSRAIYFAAY 102
> ath:AT5G48970 mitochondrial substrate carrier family protein;
K15108 solute carrier family 25 (mitochondrial thiamine pyrophosphate
transporter), member 19
Length=339
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 42/161 (26%), Positives = 75/161 (46%), Gaps = 39/161 (24%)
Query 1 FRGWLAGIVPALVLVLN-PAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTG 59
FRG+ G VPAL++V+ +IQFT+ L++ S ++ + H
Sbjct 85 FRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFA--------------SGSTKTEDH---- 126
Query 60 SLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKT----RAQSKVYNVKDYSSSV 115
I ++P SF+ G A AA L +YP+ +++T + + KVY ++
Sbjct 127 ---------IHLSPYLSFVSGALAGCAATLGSYPFDLLRTILASQGEPKVY------PTM 171
Query 116 RS-LIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
RS + I+++ G+ GL+ GL L + + + F Y+
Sbjct 172 RSAFVDIIQSRGIRGLYNGLTPTLVEIVPYAGLQFGTYDMF 212
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 41/163 (25%), Positives = 68/163 (41%), Gaps = 30/163 (18%)
Query 2 RGWLAGIVPALV-LVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
RG G+ P LV +V +QF YD + +M R++ +S+ P +
Sbjct 185 RGLYNGLTPTLVEIVPYAGLQFGTYDMFKRWMMDW-NRYKLSSKIPIN------------ 231
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKV------YNVK----D 110
+ ++ + F+ GL A +A LV +P VVK R Q + Y +
Sbjct 232 ------VDTNLSSFQLFICGLGAGTSAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRA 285
Query 111 YSSSVRSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
Y + + L I+ +EG GL+ G+ + A+ F YE
Sbjct 286 YRNMLDGLRQIMISEGWHGLYKGIVPSTVKAAPAGAVTFVAYE 328
> sce:YBR291C CTP1; Ctp1p; K15100 solute carrier family 25 (mitochondrial
citrate transporter), member 1
Length=299
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
+FL+G + + T P VKTR QS + YSS++ I + EGL + G
Sbjct 217 TFLVGAFSGIVTVYSTMPLDTVKTRMQS--LDSTKYSSTMNCFATIFKEEGLKTFWKGAT 274
Query 136 SKLTATLLSSAIMFTVYEQLL 156
+L +LS I+FT+YE++L
Sbjct 275 PRLGRLVLSGGIVFTIYEKVL 295
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 8/98 (8%)
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+S + + V P SFL G A A A +TYP+ KTR Q + S + R+ +++
Sbjct 1 MSSKATKSDVDPLHSFLAGSLAGAAEACITYPFEFAKTRLQL----IDKASKASRNPLVL 56
Query 122 L----ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
+ +T+G+ ++ G P+ + + I F ++ +
Sbjct 57 IYKTAKTQGIGSIYVGCPAFIIGNTAKAGIRFLGFDTI 94
> ath:AT4G11440 binding
Length=628
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 72 TPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLIMILETEGLSGL 130
T ++ G A AAA T P+ VVKTR Q+++ ++ SV ++L I EGL GL
Sbjct 515 TTLQTLTCGGLAGSAAAFFTTPFDVVKTRLQTQIPGSRNQHPSVYQTLQSIRRQEGLRGL 574
Query 131 FAGLPSKLTATLLSSAIMFTVYE 153
+ GL +L + AI F YE
Sbjct 575 YRGLIPRLVMYMSQGAIFFASYE 597
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 43/93 (46%), Gaps = 7/93 (7%)
Query 78 LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSK 137
G A ++ +L +P VKT QS K ++ RS+I G SGL+ G+ S
Sbjct 331 FAGALAGISVSLCLHPLDTVKTMIQSCRLEEKSLCNTGRSII---SERGFSGLYRGIASN 387
Query 138 LTATLLSSAIMFTVYE----QLLPLVVLLFRSL 166
+ ++ SA+ YE LLPL + SL
Sbjct 388 IASSAPISALYTFTYETVKGTLLPLFPKEYCSL 420
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 41/83 (49%), Gaps = 4/83 (4%)
Query 78 LMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSK 137
L G +A +A + + P +K + Q + Y + +L+ I++ GL L+AG +
Sbjct 424 LAGGSASIATSFIFTPSERIKQQMQVSSH----YRNCWTALVGIIQKGGLLSLYAGWTAV 479
Query 138 LTATLLSSAIMFTVYEQLLPLVV 160
L + S I F VYE + +V+
Sbjct 480 LCRNIPHSIIKFYVYENMKQMVL 502
> ath:AT5G64970 mitochondrial substrate carrier family protein
Length=428
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/151 (27%), Positives = 72/151 (47%), Gaps = 20/151 (13%)
Query 7 GIVPALV-LVLNPAIQFTLYDHLRAKLM---QLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G+VP+LV + + A+ + +YD L++ + + K+R +E + Q
Sbjct 282 GLVPSLVSMAPSGAVFYGVYDILKSAYLHTPEGKKRLEHMKQEGEELNAFDQ-------- 333
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMIL 122
+ + P + L G A + TYP+ VV+ R Q + + + S+V + + I+
Sbjct 334 ------LELGPMRTLLYGAIAGACSEAATYPFEVVRRRLQMQSHAKR--LSAVATCVKII 385
Query 123 ETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
E G+ L+AGL L L S+AI + VYE
Sbjct 386 EQGGVPALYAGLIPSLLQVLPSAAISYFVYE 416
> hsa:55186 SLC25A36, DKFZp564C053, FLJ10618; solute carrier family
25, member 36; K15116 solute carrier family 25, member
33/36
Length=311
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 40/165 (24%), Positives = 72/165 (43%), Gaps = 28/165 (16%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASR-EPSSNSLSSQHSSTGS 60
+G+ G+ + + I F +Y+ ++ KL++ ++TAS E S+ G
Sbjct 174 KGFYRGMSASYAGISETVIHFVIYESIKQKLLE----YKTASTMENDEESVKEASDFVGM 229
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIM 120
+ L +K A + YP+ VV+TR + + Y S ++L +
Sbjct 230 M----------------LAAATSKTCATTIAYPHEVVRTRLREEG---TKYRSFFQTLSL 270
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+++ EG L+ GL + L + ++AIM YE LVV L
Sbjct 271 LVQEEGYGSLYRGLTTHLVRQIPNTAIMMATYE----LVVYLLNG 311
Score = 33.1 bits (74), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 36/95 (37%), Gaps = 21/95 (22%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVR-------------------SLIM 120
G A++T P VVKTR QS +V Y S V+ L +
Sbjct 13 GGCGGTVGAILTCPLEVVKTRLQSS--SVTLYISEVQLNTMAGASVNRVVSPGPLHCLKV 70
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
ILE EG LF GL L S AI F Y
Sbjct 71 ILEKEGPRSLFRGLGPNLVGVAPSRAIYFAAYSNC 105
> sce:YPR021C AGC1; Agc1p; K15105 solute carrier family 25 (mitochondrial
aspartate/glutamate transporter), member 12/13
Length=902
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLP 135
+F +G A A V YP +KTR Q++ ++ Y +S+ L+ I+ EG+ GL++GL
Sbjct 533 NFSLGSIAGCIGATVVYPIDFIKTRMQAQ-RSLAQYKNSIDCLLKIISREGIKGLYSGLG 591
Query 136 SKLTATLLSSAIMFTV 151
+L AI TV
Sbjct 592 PQLIGVAPEKAIKLTV 607
Score = 31.6 bits (70), Expect = 1.5, Method: Composition-based stats.
Identities = 35/156 (22%), Positives = 63/156 (40%), Gaps = 34/156 (21%)
Query 2 RGWLAGIVPALVLVL-NPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGS 60
+G +G+ P L+ V AI+ T+ D +R +L
Sbjct 584 KGLYSGLGPQLIGVAPEKAIKLTVNDFMRNRL---------------------------- 615
Query 61 LVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKV-YNVKDYSSSVRSLI 119
+D+ ++ + P + G +A + T P +VK R Q + Y ++ + +
Sbjct 616 --TDKNGKLSLFP--EIISGASAGACQVIFTNPLEIVKIRLQVQSDYVGENIQQANETAT 671
Query 120 MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
I++ GL GL+ G+ + L + SAI F Y L
Sbjct 672 QIVKKLGLRGLYNGVAACLMRDVPFSAIYFPTYAHL 707
> cel:C16C10.1 hypothetical protein; K15119 solute carrier family
25, member 39/40
Length=360
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 47/80 (58%), Gaps = 2/80 (2%)
Query 76 SFLMGLAAKLAAALVTYPYLVVKTRAQSKV-YNVKDYSSSVRSLIM-ILETEGLSGLFAG 133
SF+ G AA + A++ T+P+ V+KT Q ++ ++ D + S+ ++I + + G+S +G
Sbjct 271 SFVSGAAAGVVASIFTHPFDVIKTNCQIRIGGSIDDMNKSITTVIKDMYHSRGISAFSSG 330
Query 134 LPSKLTATLLSSAIMFTVYE 153
L +L S AIM + YE
Sbjct 331 LVPRLVKVSPSCAIMISFYE 350
> sce:YFR045W Putative mitochondrial transport protein; null mutant
is viable, exhibits decreased levels of chitin and normal
resistance to calcofluor white
Length=309
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 47/92 (51%), Gaps = 3/92 (3%)
Query 65 RMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVK-DYSSSVRSLIMILE 123
R+L+ R A S + GLA +T P VVKTR S+ N K +Y +++ + I
Sbjct 210 RLLQARNDKASSVITGLATSFTLVAMTQPIDVVKTRMMSQ--NAKTEYKNTLNCMYRIFV 267
Query 124 TEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG++ + G + +S + FTVYEQ+
Sbjct 268 QEGMATFWKGSIFRFMKVGISGGLTFTVYEQV 299
> sce:YGR096W TPC1; Mitochondrial membrane transporter that mediates
uptake of the essential cofactor thiamine pyrophosphate
(ThPP) into mitochondria; expression appears to be regulated
by carbon source; member of the mitochondrial carrier family;
K15108 solute carrier family 25 (mitochondrial thiamine
pyrophosphate transporter), member 19
Length=314
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 49/82 (59%), Gaps = 4/82 (4%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV-RSLIMILETEGLSGLFAG 133
S ++G A + +++V+YP+ V++TR V N + +S S+ R + I + EGL G F G
Sbjct 114 HSLVVGAFAGITSSIVSYPFDVLRTRL---VANNQMHSMSITREVRDIWKLEGLPGFFKG 170
Query 134 LPSKLTATLLSSAIMFTVYEQL 155
+ +T L+++IMF YE +
Sbjct 171 SIASMTTITLTASIMFGTYETI 192
Score = 29.6 bits (65), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 13/87 (14%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQ-------------SKVYNVKDYSSSVRSLIMILETEG 126
G + A ++T+P ++ R Q S VY R + IL+ EG
Sbjct 219 GTIGGVIAKIITFPLETIRRRMQFMNSKHLEKFSRHSSVYGSYKGYGFARIGLQILKQEG 278
Query 127 LSGLFAGLPSKLTATLLSSAIMFTVYE 153
+S L+ G+ L+ T+ ++ + F YE
Sbjct 279 VSSLYRGILVALSKTIPTTFVSFWGYE 305
> hsa:81034 SLC25A32, FLJ23872, MFT, MFTC; solute carrier family
25, member 32; K15115 solute carrier family 25 (mitochondrial
folate transporter), member 32
Length=315
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 60/154 (38%), Gaps = 30/154 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG G VP L + A+QF Y+ L+ K Q +R P + + ++ S +L
Sbjct 180 RGLYKGFVPGLFGTSHGALQFMAYELLKLKYNQ------HINRLPEAQLSTVEYISVAAL 233
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+K+ A TYPY VV+ R Q + YS + +
Sbjct 234 ---------------------SKIFAVAATYPYQVVRARLQDQ---HMFYSGVIDVITKT 269
Query 122 LETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ G + G+ L + I F VYE +
Sbjct 270 WRKEGVGGFYKGIAPNLIRVTPACCITFVVYENV 303
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + G++ + + L +P +VK R A S ++ Y+ + L I + +GL GL+
Sbjct 24 ENLIAGVSGGVLSNLALHPLDLVKIRFAVSDGLELRPKYNGILHCLTTIWKLDGLRGLYQ 83
Query 133 GLPSKLTATLLSSAIMFTVYEQL 155
G+ + LS + F Y +
Sbjct 84 GVTPNIWGAGLSWGLYFFFYNAI 106
Score = 28.9 bits (63), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 36/91 (39%), Gaps = 6/91 (6%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV-----KDYSSSVRSLIMILET 124
R+ E + A +T P V KTR + V + Y +L+ I +
Sbjct 117 RLEATEYLVSAAEAGAMTLCITNPLWVTKTRLMLQYDAVVNSPHRQYKGMFDTLVKIYKY 176
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ GL+ G L T A+ F YE L
Sbjct 177 EGVRGLYKGFVPGLFGT-SHGALQFMAYELL 206
> ath:AT4G39460 SAMC1; SAMC1 (S-ADENOSYLMETHIONINE CARRIER 1);
S-adenosylmethionine transmembrane transporter/ binding; K15111
solute carrier family 25 (mitochondrial S-adenosylmethionine
transporter), member 26
Length=325
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 40/77 (51%), Gaps = 5/77 (6%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGLPSKLT 139
G LAA+L+ P VVK R Q+ ++S+ ++ MI EG GL+AG S L
Sbjct 142 GAIGGLAASLIRVPTEVVKQRMQTG-----QFTSAPSAVRMIASKEGFRGLYAGYRSFLL 196
Query 140 ATLLSSAIMFTVYEQLL 156
L AI F +YEQL
Sbjct 197 RDLPFDAIQFCIYEQLC 213
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 40/154 (25%), Positives = 59/154 (38%), Gaps = 33/154 (21%)
Query 1 FRGWLAGIVPALVLVLN-PAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTG 59
FRG AG L+ L AIQF +Y+ QL ++ A+R S+
Sbjct 184 FRGLYAGYRSFLLRDLPFDAIQFCIYE-------QLCLGYKKAARRELSD---------- 226
Query 60 SLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLI 119
P + + A L A VT P V+KTR + + K Y V +
Sbjct 227 -------------PENALIGAFAGALTGA-VTTPLDVIKTRLMVQ-GSAKQYQGIVDCVQ 271
Query 120 MILETEGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
I+ EG L G+ ++ + +I F V E
Sbjct 272 TIVREEGAPALLKGIGPRVLWIGIGGSIFFGVLE 305
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 33/79 (41%), Gaps = 15/79 (18%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
E F+ G A + YP +KTR Q+ R I+ L GL++GL
Sbjct 56 EGFIAGGTAGVVVETALYPIDTIKTRLQA-----------ARGGGKIV----LKGLYSGL 100
Query 135 PSKLTATLLSSAIMFTVYE 153
+ L +SA+ VYE
Sbjct 101 AGNIAGVLPASALFVGVYE 119
> dre:503758 slc25a32b, zgc:110786; solute carrier family 25,
member 32b; K15115 solute carrier family 25 (mitochondrial folate
transporter), member 32
Length=313
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 57/149 (38%), Gaps = 30/149 (20%)
Query 7 GIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLVSDRM 66
G VP L + A+QF Y+ L+ + +++ A
Sbjct 186 GFVPGLFGTSHGALQFMAYEELKRDYNKYRKKQSDA------------------------ 221
Query 67 LRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEG 126
++ P E M +K+ A TYPY VV+ R Q + +N Y+ + EG
Sbjct 222 ---KLNPLEYITMAALSKIFAVATTYPYQVVRARLQDQ-HNT--YNGLTDVVWRTWRNEG 275
Query 127 LSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
L G + G+ L + I F VYE +
Sbjct 276 LLGFYKGMVPNLVRVTPACCITFVVYENV 304
Score = 38.1 bits (87), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 40/91 (43%), Gaps = 4/91 (4%)
Query 68 RIRVTPAESFLMGLAAKLAAALVTYPYLVVKTR---AQSKVYNVKDYSSSVRSLIMILET 124
+I +T E L A +T P V KTR S + K Y + +L+ I
Sbjct 118 QIELTATEHLLSAAVAGAMTLCLTNPIWVTKTRLVLQYSADPSQKQYKGMMDALVKIYRH 177
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+SGL+ G L T A+ F YE+L
Sbjct 178 EGISGLYRGFVPGLFGT-SHGALQFMAYEEL 207
Score = 33.5 bits (75), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + GL+ + + L +P +VK R A S +V+ YS V + I EG GL+
Sbjct 27 ENLIAGLSGGVLSTLALHPLDLVKIRFAVSDGLDVRPKYSGIVHCMKSIWHQEGFRGLYQ 86
Query 133 GLPSKLTATLLSSAIMFTVYEQL 155
G+ + S + F Y +
Sbjct 87 GVTPNIWGAGASWGLYFFFYNAI 109
> hsa:54977 SLC25A38, FLJ20551, FLJ22703; solute carrier family
25, member 38; K15118 solute carrier family 25, member 38
Length=304
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 51/97 (52%), Gaps = 10/97 (10%)
Query 59 GSLVSDRMLRIRVTPA---ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV 115
G+L S + +R P ES ++G+ ++ A + P V+KTR +S Y + +++
Sbjct 106 GTLYSLKQYFLRGHPPTALESVMLGVGSRSVAGVCMSPITVIKTRYESGKYGYESIYAAL 165
Query 116 RSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVY 152
RS I +EG GLF+G LTATLL A +Y
Sbjct 166 RS---IYHSEGHRGLFSG----LTATLLRDAPFSGIY 195
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/103 (31%), Positives = 59/103 (57%), Gaps = 5/103 (4%)
Query 55 HSSTGSLVSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSS 114
++ T ++V + + P +F G+ A + A+LVT P V+KT Q +Y +K +
Sbjct 199 YNQTKNIVPHDQVDATLIPITNFSCGIFAGILASLVTQPADVIKTHMQ--LYPLK-FQWI 255
Query 115 VRSLIMILETEGLSGLF-AGLPSKLTATLLSSAIMFTVYEQLL 156
+++ +I + GL G F G+P L TL+ +A+ +TVYE+++
Sbjct 256 GQAVTLIFKDYGLRGFFQGGIPRALRRTLM-AAMAWTVYEEMM 297
> ath:AT3G55640 mitochondrial substrate carrier family protein
Length=332
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 44/81 (54%), Gaps = 5/81 (6%)
Query 77 FLMGLAAKLAAALVTYPYLVVKTR--AQSKVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
F+ G A + AA TYP +V+TR AQ+KV YS +L I EG+ GL+ GL
Sbjct 145 FVAGGLAGITAASATYPLDLVRTRLAAQTKVIY---YSGIWHTLRSITTDEGILGLYKGL 201
Query 135 PSKLTATLLSSAIMFTVYEQL 155
+ L S AI F+VYE L
Sbjct 202 GTTLVGVGPSIAISFSVYESL 222
> mmu:69906 Slc25a32, 2610043O12Rik, Mftc; solute carrier family
25, member 32; K15115 solute carrier family 25 (mitochondrial
folate transporter), member 32
Length=316
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/154 (24%), Positives = 62/154 (40%), Gaps = 30/154 (19%)
Query 2 RGWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSL 61
RG G VP L + A+QF Y+ L++LK + +R P + ++++ S +L
Sbjct 180 RGLYKGFVPGLFGTSHGALQFMAYE-----LLKLKYN-KHINRLPEAQLSTAEYISVAAL 233
Query 62 VSDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMI 121
+K+ A TYPY VV+ R Q + + Y +
Sbjct 234 ---------------------SKIFAVAATYPYQVVRARLQDQHVS---YGGVTDVITKT 269
Query 122 LETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ G + G+ L + I F VYE +
Sbjct 270 WRKEGIGGFYKGIAPNLIRVTPACCITFVVYENV 303
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 28/102 (27%), Positives = 45/102 (44%), Gaps = 14/102 (13%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + G++ + + L +P +VK R A S V+ Y + L I + +GL GL+
Sbjct 24 ENLVAGVSGGVLSNLALHPLDLVKIRFAVSDGLEVRPKYKGILHCLATIWKVDGLRGLYQ 83
Query 133 GLPSKLTATLLSSAIMFTVY------------EQLLPLVVLL 162
G+ + LS + F Y EQL PL L+
Sbjct 84 GVTPNVWGAGLSWGLYFFFYNAIKSYKTEGRAEQLEPLEYLV 125
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 37/91 (40%), Gaps = 6/91 (6%)
Query 70 RVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNV-----KDYSSSVRSLIMILET 124
++ P E + A +T P V KTR + V + Y +L+ I +
Sbjct 117 QLEPLEYLVSAAEAGAMTLCITNPLWVTKTRLMLQYGGVASPSQRQYKGMFDALVKIYKY 176
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYEQL 155
EG+ GL+ G L T A+ F YE L
Sbjct 177 EGVRGLYKGFVPGLFGT-SHGALQFMAYELL 206
> mmu:208638 Slc25a38, AV019094, BC010801, MGC18873; solute carrier
family 25, member 38; K15118 solute carrier family 25,
member 38
Length=326
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 51/97 (52%), Gaps = 10/97 (10%)
Query 59 GSLVSDRMLRIRVTPA---ESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSV 115
G+L S + +R P ES ++G+ ++ A + P V+KTR +S Y+ + +++
Sbjct 126 GTLYSSKQYFLRGHPPTALESVILGMGSRSVAGVCMSPITVIKTRYESGTYSYESIYAAL 185
Query 116 RSLIMILETEGLSGLFAGLPSKLTATLLSSAIMFTVY 152
RS I +EG GLF G LTATLL A +Y
Sbjct 186 RS---IYCSEGHRGLFRG----LTATLLRDAPFSGLY 215
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 49/85 (57%), Gaps = 5/85 (5%)
Query 73 PAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFA 132
P +F G+ A + A+LVT P V+KT Q + VK + ++ +I + GL G F
Sbjct 239 PLINFSCGIFAGVLASLVTQPADVIKTHMQ--LSPVK-FQWIGQAATLIFKNHGLRGFFH 295
Query 133 G-LPSKLTATLLSSAIMFTVYEQLL 156
G +P L TL++ A+ +TVYE+++
Sbjct 296 GSVPRALRRTLMA-AMAWTVYEEMM 319
> dre:100330582 hypothetical protein LOC100330582; K15100 solute
carrier family 25 (mitochondrial citrate transporter), member
1
Length=317
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 47/95 (49%), Gaps = 2/95 (2%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGL 130
+ P + L G A A+ P V+KTR Q Y S+V I I++ EG +
Sbjct 224 INPVVTGLFGAVAGAASVFGNTPLDVIKTRMQG--LEAHKYKSTVDCAIKIMKYEGPAAF 281
Query 131 FAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+ G +L + AI+F +YE+++ ++ ++++
Sbjct 282 YKGTVPRLGRVCMDVAIVFIIYEEVVKVLNKVWKT 316
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 1/84 (1%)
Query 73 PAESFLMGLAAKLAAALVTYPYLVVKTRAQ-SKVYNVKDYSSSVRSLIMILETEGLSGLF 131
P ++ L G A +T+P VKT+ Q + N Y V + ++ G+ GL+
Sbjct 31 PGKAILAGGIAGGIEICITFPTEYVKTQLQLDEKANPPRYKGIVDCVKQTVQGHGVKGLY 90
Query 132 AGLPSKLTATLLSSAIMFTVYEQL 155
GL S L ++ +A+ F V+E L
Sbjct 91 RGLSSLLYGSIPKAAVRFGVFEFL 114
> dre:795332 similar to Tricarboxylate transport protein, mitochondrial
precursor (Citrate transport protein) (CTP) (Tricarboxylate
carrier protein) (Solute carrier family 25 member
1); K15100 solute carrier family 25 (mitochondrial citrate transporter),
member 1
Length=317
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 47/95 (49%), Gaps = 2/95 (2%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGL 130
+ P + L G A A+ P V+KTR Q Y S+V I I++ EG +
Sbjct 224 INPVVTGLFGAVAGAASVFGNTPLDVIKTRMQG--LEAHKYKSTVDCAIKIMKYEGPAAF 281
Query 131 FAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRS 165
+ G +L + AI+F +YE+++ ++ ++++
Sbjct 282 YKGTVPRLGRVCMDVAIVFIIYEEVVKVLNKVWKT 316
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 1/84 (1%)
Query 73 PAESFLMGLAAKLAAALVTYPYLVVKTRAQ-SKVYNVKDYSSSVRSLIMILETEGLSGLF 131
P ++ L G A +T+P VKT+ Q + N Y V + ++ G+ GL+
Sbjct 31 PGKAILAGGIAGGIEICITFPTEYVKTQLQLDEKANPPRYKGIVDCVKQTVQGHGVKGLY 90
Query 132 AGLPSKLTATLLSSAIMFTVYEQL 155
GL S L ++ +A+ F V+E L
Sbjct 91 RGLSSLLYGSIPKAAVRFGVFEFL 114
> hsa:123096 SLC25A29, C14orf69, CACL, FLJ38975; solute carrier
family 25, member 29; K15109 solute carrier family 25 (mitochondrial
carnitine/acylcarnitine transporter), member 20/29
Length=303
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 39/83 (46%), Gaps = 0/83 (0%)
Query 74 AESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAG 133
A FL G A +A LV +P+ VK R Q + Y ++ I++ E + GL+ G
Sbjct 2 ALDFLAGCAGGVAGVLVGHPFDTVKVRLQVQSVEKPQYRGTLHCFKSIIKQESVLGLYKG 61
Query 134 LPSKLTATLLSSAIMFTVYEQLL 156
L S L +A++F V L
Sbjct 62 LGSPLMGLTFINALVFGVQGNTL 84
> mmu:214663 Slc25a29, C030003J19Rik, CACL, mCACL; solute carrier
family 25 (mitochondrial carrier, palmitoylcarnitine transporter),
member 29; K15109 solute carrier family 25 (mitochondrial
carnitine/acylcarnitine transporter), member 20/29
Length=306
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 74 AESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAG 133
A FL G A +A +V +P+ +VK R Q + Y ++ I++ E + GL+ G
Sbjct 2 ALDFLAGCAGGVAGVIVGHPFDIVKVRLQVQSTEKPQYRGTLHCFQSIIKQESVLGLYKG 61
Query 134 LPSKLTATLLSSAIMFTVYEQLL 156
L S L +A++F V L
Sbjct 62 LGSPLMGLTFINALVFGVQGNTL 84
> dre:571961 slc25a38b; Solute carrier family 25 member 38-B;
K15118 solute carrier family 25, member 38
Length=287
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 41/79 (51%), Gaps = 7/79 (8%)
Query 74 AESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGLFAG 133
E+ L+G A+ A + P+ V+KTR +S YN Y S +L + + EG L++G
Sbjct 107 GEAVLLGAGARCVAGVAMLPFTVIKTRFESGRYN---YISVAGALKSVCQNEGPKALYSG 163
Query 134 LPSKLTATLLSSAIMFTVY 152
LTATLL A +Y
Sbjct 164 ----LTATLLRDAPFSGIY 178
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 47/87 (54%), Gaps = 5/87 (5%)
Query 71 VTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILETEGLSGL 130
+ P +F G+ A + A+L T P V+KT Q S ++R + + GLSG
Sbjct 198 IAPLVNFGCGVVAGILASLATQPADVIKTHMQVSPALYPKTSDAMRHVYV---KHGLSGF 254
Query 131 FAG-LPSKLTATLLSSAIMFTVYEQLL 156
F G +P L TL++ A+ +TVYEQL+
Sbjct 255 FRGAVPRSLRRTLMA-AMAWTVYEQLM 280
> cpv:cgd1_2370 mitochondrial carrier protein, Flx1p like mitochondrial
membrane associated flavin transporter with 4 or more
transmembrane domains
Length=329
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/162 (23%), Positives = 72/162 (44%), Gaps = 37/162 (22%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G +G +P L+L+ + IQ +YD R + S + +S+H
Sbjct 191 GLYSGFIPTLMLIPHTLIQLVIYDIFRNQ----------------SINYNSKH------- 227
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQ-SKVYNVKDYSSSVRSLIMI 121
+ ++ F+ G +KL A+ +TYP V+++R Q +K+ N++ + + I+
Sbjct 228 ------LYLSNLRFFIFGFISKLLASALTYPLQVIRSRMQMAKIENIQLKTYDIN--ILK 279
Query 122 LETEGLSGL-----FAGLPSKLTATLLSSAIMFTVYEQLLPL 158
L + F GL + + + S IMF +YE ++ L
Sbjct 280 LSHKEFVNFARDHYFPGLMTHIPKVSMHSGIMFLIYEAMIKL 321
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 42/75 (56%), Gaps = 7/75 (9%)
Query 77 FLMGLAAKLAAALVT----YPYLVVKTRAQSKVYN---VKDYSSSVRSLIMILETEGLSG 129
F + + L +A++T +P VV+TR Q + V + + + ++ I++TEG+ G
Sbjct 12 FFLTATSSLISAIITSFLLHPLDVVRTRQQVAAASDGAVIAHPTLISTINFIIDTEGVIG 71
Query 130 LFAGLPSKLTATLLS 144
L+ GL +L A+ +S
Sbjct 72 LYKGLNGQLVASGVS 86
> xla:495984 slc25a32; solute carrier family 25, member 32; K15115
solute carrier family 25 (mitochondrial folate transporter),
member 32
Length=318
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 65/165 (39%), Gaps = 31/165 (18%)
Query 3 GWLAGIVPALVLVLNPAIQFTLYDHLRAKLMQLKQRWRTASREPSSNSLSSQHSSTGSLV 62
G G VP L+ + A+QF Y+ L+ + + R PS L G+L
Sbjct 184 GLYKGFVPGLLGTSHGALQFMAYEELKMEYNKYLNR-------PSDTKL-------GTL- 228
Query 63 SDRMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMIL 122
E M +K+ A TYPY VV+ R Q + Y+ + +
Sbjct 229 ------------EYITMAALSKIFAVSTTYPYQVVRARLQDQ---HNRYTGVLDVISRTW 273
Query 123 ETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLR 167
EG+ G + G+ + + I F VYE++ +L FR R
Sbjct 274 RKEGVQGFYKGIVPNIIRVTPACCITFVVYEKVSHF-LLDFRKHR 317
Score = 32.7 bits (73), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 38/81 (46%), Gaps = 2/81 (2%)
Query 75 ESFLMGLAAKLAAALVTYPYLVVKTR-AQSKVYNVK-DYSSSVRSLIMILETEGLSGLFA 132
E+ + GL+ + + LV +P +VK R A S ++ Y V L + + EGL GL+
Sbjct 27 ENLVAGLSGGVISTLVLHPLDLVKIRFAVSDGLELRPKYRGIVHCLATVWQREGLRGLYQ 86
Query 133 GLPSKLTATLLSSAIMFTVYE 153
G+ + S + F Y
Sbjct 87 GVTPNMWGAGASWGLYFFFYN 107
> ath:AT1G78180 binding
Length=418
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 72/149 (48%), Gaps = 16/149 (10%)
Query 7 GIVPALV-LVLNPAIQFTLYDHLRAKLMQLKQ-RWRTASREPSSNSLSSQHSSTGSLVSD 64
G+VP++ + L+ A+ + +YD L++ + + R R + L++
Sbjct 275 GLVPSIASMALSGAVFYGVYDILKSSFLHTPEGRKRLIDMKQQGQELNA----------- 323
Query 65 RMLRIRVTPAESFLMGLAAKLAAALVTYPYLVVKTRAQSKVYNVKDYSSSVRSLIMILET 124
+ R+ + P + + G A + TYP+ VV+ + Q ++ K+ +++ I+E
Sbjct 324 -LDRLELGPIRTLMYGAIAGACTEVATYPFEVVRRQLQMQM--GKNKLNALAMGFNIIER 380
Query 125 EGLSGLFAGLPSKLTATLLSSAIMFTVYE 153
G+ L+AGL L L S++I + VYE
Sbjct 381 GGIPALYAGLLPSLLQVLPSASISYFVYE 409
Score = 30.4 bits (67), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 121 ILETEGLSGLFAGLPSKLTATLLSSAIMFTVYEQLLPLVVLLFRSLRVPTYRKR 174
+++TEGL L+ GL + + LS A+ + VY+ +L L P RKR
Sbjct 263 MIQTEGLFSLYKGLVPSIASMALSGAVFYGVYD------ILKSSFLHTPEGRKR 310
> sce:YKL120W OAC1; Mitochondrial inner membrane transporter,
transports oxaloacetate, sulfate, thiosulfate, and isopropylmalate;
member of the mitochondrial carrier family; K15117 solute
carrier family 25, member 34/35
Length=324
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 80 GLAAKLAAALVTYPYLVVKTRAQS-----KVYNVKDYSSSVRSLIMILETEGLSGLFAGL 134
G A+ + A++ P +VKTR QS K+ Y+ L+ I +TEG+ GLF G+
Sbjct 135 GAASGIIGAVIGSPLFLVKTRLQSYSEFIKIGEQTHYTGVWNGLVTIFKTEGVKGLFRGI 194
Query 135 PSKLTATLLSSAIMFTVYE 153
+ + T S++ +Y
Sbjct 195 DAAILRTGAGSSVQLPIYN 213
Lambda K H
0.322 0.133 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40