bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0305_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
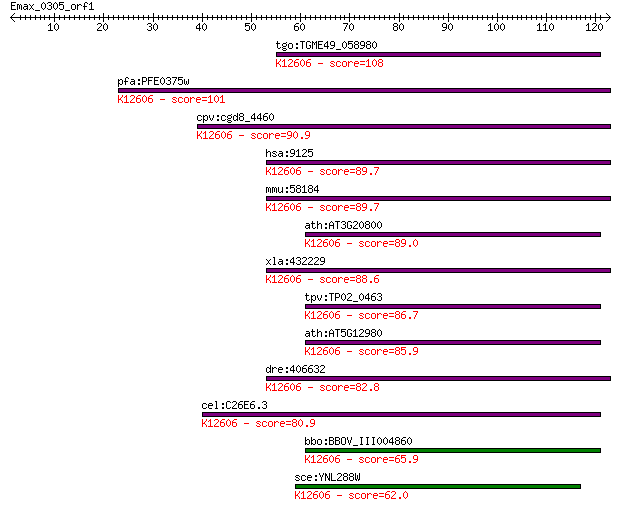
tgo:TGME49_058980 cell differentiation protein Rcd1, putative ... 108 3e-24
pfa:PFE0375w cell differentiation protein rcd1, putative; K126... 101 5e-22
cpv:cgd8_4460 cell differentiation protein rcd1 ; K12606 CCR4-... 90.9 8e-19
hsa:9125 RQCD1, CNOT9, CT129, RCD1, RCD1+; RCD1 required for c... 89.7 2e-18
mmu:58184 Rqcd1, 2610007F23Rik, AI593551, Fl10; rcd1 (required... 89.7 2e-18
ath:AT3G20800 rcd1-like cell differentiation protein, putative... 89.0 3e-18
xla:432229 rqcd1, MGC78923, rcd1; RCD1 required for cell diffe... 88.6 4e-18
tpv:TP02_0463 hypothetical protein; K12606 CCR4-NOT transcript... 86.7 2e-17
ath:AT5G12980 rcd1-like cell differentiation protein, putative... 85.9 3e-17
dre:406632 rqcd1, wu:fb52b11, wu:fb74e08, zgc:85618; RCD1 requ... 82.8 3e-16
cel:C26E6.3 hypothetical protein; K12606 CCR4-NOT transcriptio... 80.9 9e-16
bbo:BBOV_III004860 17.m07435; cell differentiation family prot... 65.9 3e-11
sce:YNL288W CAF40; Caf40p; K12606 CCR4-NOT transcription compl... 62.0 5e-10
> tgo:TGME49_058980 cell differentiation protein Rcd1, putative
; K12606 CCR4-NOT transcription complex subunit 9
Length=356
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 47/66 (71%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 55 QQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFL 114
+D +CQL+LDL + EKRE AL +LSKRRE +PDLAPLLW SFGT+AA+LQEIIA YP L
Sbjct 78 DKDHVCQLLLDLCVLEKREAALADLSKRREQYPDLAPLLWHSFGTIAAILQEIIAIYPCL 137
Query 115 SPPSLT 120
SPP+LT
Sbjct 138 SPPTLT 143
> pfa:PFE0375w cell differentiation protein rcd1, putative; K12606
CCR4-NOT transcription complex subunit 9
Length=652
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 43/100 (43%), Positives = 63/100 (63%), Gaps = 0/100 (0%)
Query 23 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQDRICQLVLDLSIAEKREGALLELSKR 82
+ +Q Q + Q+ ++++ ++ QLV DL +EKRE ALLELS++
Sbjct 193 KDNTTSEQDQTKNDNNINNLSSGQENTINNEEEKKKVYQLVYDLCFSEKRENALLELSRK 252
Query 83 RESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLTTQ 122
RE++ D+AP+LW SFGT+ LLQEI++ YP LSPP LTT
Sbjct 253 RETYHDIAPVLWNSFGTITTLLQEIVSIYPQLSPPLLTTS 292
> cpv:cgd8_4460 cell differentiation protein rcd1 ; K12606 CCR4-NOT
transcription complex subunit 9
Length=390
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 59/84 (70%), Gaps = 0/84 (0%)
Query 39 QQQQQQQQQQQQQQQQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFG 98
+ + Q++++++ Q ++D++ + KRE AL +LSK RE+FPDLAPL+W SFG
Sbjct 108 SDNSGDLKGNENIIQKEREKVYQYLVDMTCSSKREMALSKLSKYRETFPDLAPLMWHSFG 167
Query 99 TMAALLQEIIAAYPFLSPPSLTTQ 122
+ ALLQEII+ YP LSPP+L+ Q
Sbjct 168 CITALLQEIISIYPLLSPPNLSNQ 191
> hsa:9125 RQCD1, CNOT9, CT129, RCD1, RCD1+; RCD1 required for
cell differentiation1 homolog (S. pombe); K12606 CCR4-NOT transcription
complex subunit 9
Length=299
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/70 (60%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 53 QQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYP 112
Q +++I Q + +LS E RE ALLELSK+RES PDLAP+LW SFGT+AALLQEI+ YP
Sbjct 16 QVDREKIYQWINELSSPETRENALLELSKKRESVPDLAPMLWHSFGTIAALLQEIVNIYP 75
Query 113 FLSPPSLTTQ 122
++PP+LT
Sbjct 76 SINPPTLTAH 85
> mmu:58184 Rqcd1, 2610007F23Rik, AI593551, Fl10; rcd1 (required
for cell differentiation) homolog 1 (S. pombe); K12606 CCR4-NOT
transcription complex subunit 9
Length=299
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/70 (60%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 53 QQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYP 112
Q +++I Q + +LS E RE ALLELSK+RES PDLAP+LW SFGT+AALLQEI+ YP
Sbjct 16 QVDREKIYQWINELSSPETRENALLELSKKRESVPDLAPMLWHSFGTIAALLQEIVNIYP 75
Query 113 FLSPPSLTTQ 122
++PP+LT
Sbjct 76 SINPPTLTAH 85
> ath:AT3G20800 rcd1-like cell differentiation protein, putative;
K12606 CCR4-NOT transcription complex subunit 9
Length=316
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 44/60 (73%), Positives = 50/60 (83%), Gaps = 0/60 (0%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QLVLDLS E RE ALLELSK+RE F DLAPLLW SFGT+AALLQEI++ Y L+PP+LT
Sbjct 40 QLVLDLSNPELRENALLELSKKRELFQDLAPLLWNSFGTIAALLQEIVSIYSVLAPPNLT 99
> xla:432229 rqcd1, MGC78923, rcd1; RCD1 required for cell differentiation1
homolog; K12606 CCR4-NOT transcription complex
subunit 9
Length=299
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 41/70 (58%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 53 QQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYP 112
Q +++I Q + +LS + RE ALLELSK+RES PDLAP+LW SFGT+AALLQEI+ YP
Sbjct 16 QVDREKIYQWINELSSPDTRENALLELSKKRESVPDLAPMLWHSFGTIAALLQEIVNIYP 75
Query 113 FLSPPSLTTQ 122
++PP+LT
Sbjct 76 SINPPTLTAH 85
> tpv:TP02_0463 hypothetical protein; K12606 CCR4-NOT transcription
complex subunit 9
Length=327
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/60 (63%), Positives = 50/60 (83%), Gaps = 0/60 (0%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QL+LDLSI EKRE AL+ELSK+RE++PDLA LLW SFGT+A LL EI++ Y +L P +++
Sbjct 57 QLILDLSIPEKREYALIELSKQRENYPDLAILLWHSFGTVATLLYEIVSVYHYLYPLTIS 116
> ath:AT5G12980 rcd1-like cell differentiation protein, putative;
K12606 CCR4-NOT transcription complex subunit 9
Length=311
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/60 (71%), Positives = 48/60 (80%), Gaps = 0/60 (0%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QL+LDLS E RE AL ELSK+RE F DLAPLLW S GT+ ALLQEIIA YP LSPP++T
Sbjct 36 QLILDLSNPELRENALHELSKKREIFQDLAPLLWHSVGTIPALLQEIIAVYPALSPPTMT 95
> dre:406632 rqcd1, wu:fb52b11, wu:fb74e08, zgc:85618; RCD1 required
for cell differentiation1 homolog (S. pombe); K12606
CCR4-NOT transcription complex subunit 9
Length=298
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 52/70 (74%), Gaps = 0/70 (0%)
Query 53 QQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYP 112
Q +++I Q + +LS E RE ALLELSK+RES DLAP+LW S GT+AALLQEI+ YP
Sbjct 15 QVDREKIYQWINELSSPETRENALLELSKKRESVTDLAPMLWHSCGTIAALLQEIVNIYP 74
Query 113 FLSPPSLTTQ 122
++PP+LT
Sbjct 75 SINPPTLTAH 84
> cel:C26E6.3 hypothetical protein; K12606 CCR4-NOT transcription
complex subunit 9
Length=321
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 41/81 (50%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 40 QQQQQQQQQQQQQQQQQDRICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGT 99
Q QQ D I Q ++DL KRE ALLELSK+R+S PDL LW SFGT
Sbjct 11 QAQQTAVPSSANLDINTDEIMQWIIDLRDPPKREAALLELSKKRDSVPDLPIWLWHSFGT 70
Query 100 MAALLQEIIAAYPFLSPPSLT 120
M+ALLQE++A YP + P +LT
Sbjct 71 MSALLQEVVAIYPAIMPANLT 91
> bbo:BBOV_III004860 17.m07435; cell differentiation family protein;
K12606 CCR4-NOT transcription complex subunit 9
Length=311
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/60 (56%), Positives = 41/60 (68%), Gaps = 7/60 (11%)
Query 61 QLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSPPSLT 120
QL+LDL++ EKRE AL ELSK+RE PDL LLW+SFGTMA L +L P SL+
Sbjct 47 QLILDLAVPEKREYALAELSKQREHHPDLPVLLWYSFGTMATLFH-------YLHPMSLS 99
> sce:YNL288W CAF40; Caf40p; K12606 CCR4-NOT transcription complex
subunit 9
Length=373
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/58 (53%), Positives = 39/58 (67%), Gaps = 4/58 (6%)
Query 59 ICQLVLDLSIAEKREGALLELSKRRESFPDLAPLLWFSFGTMAALLQEIIAAYPFLSP 116
ICQL + ++E ALLEL ++RE F DLA +LW SFG M +LL EII+ YP L P
Sbjct 110 ICQL----TYGPQKEQALLELGRKREQFDDLAVVLWSSFGVMTSLLNEIISVYPMLQP 163
Lambda K H
0.312 0.121 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40