bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0420_orf1
Length=145
Score E
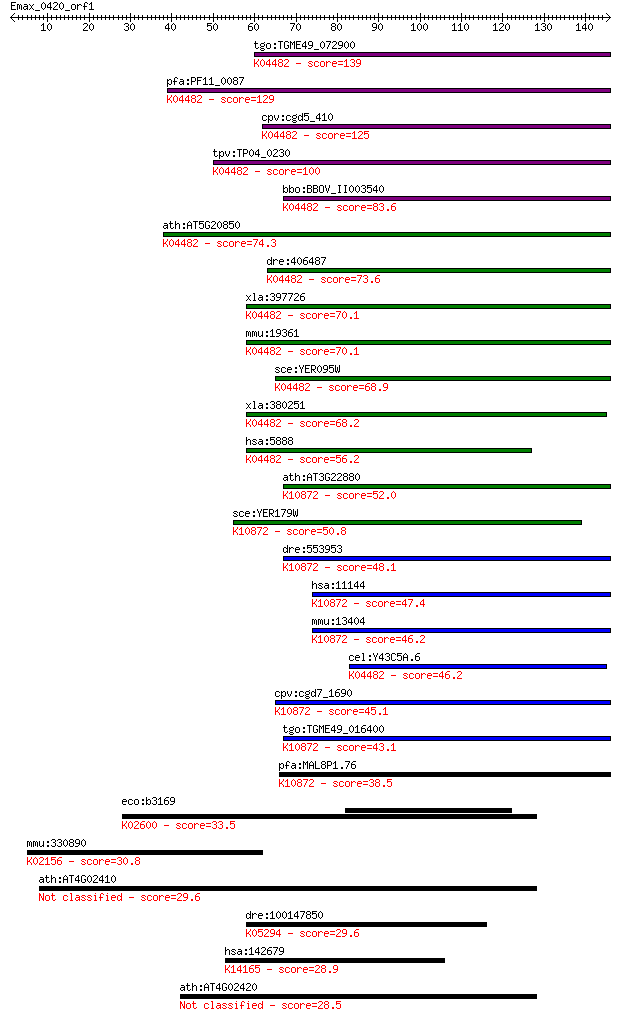
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8); K... 139 4e-33
pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein ... 129 4e-30
cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51 125 6e-29
tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair prot... 100 2e-21
bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair ... 83.6 2e-16
ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-depende... 74.3 1e-13
dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA h... 73.6 2e-13
xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad... 70.1 2e-12
mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cer... 70.1 2e-12
sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51 68.9 4e-12
xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,... 68.2 8e-12
hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA... 56.2 3e-08
ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1); AT... 52.0 6e-07
sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination pr... 50.8 1e-06
dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor... 48.1 1e-05
hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473, d... 47.4 1e-05
mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage sup... 46.2 3e-05
cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-... 46.2 3e-05
cpv:cgd7_1690 meiotic recombination protein DMC1-like protein ... 45.1 8e-05
tgo:TGME49_016400 meiotic recombination protein DMC1-like prot... 43.1 3e-04
pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;... 38.5 0.007
eco:b3169 nusA, ECK3158, JW3138; transcription termination/ant... 33.5 0.25
mmu:330890 Piwil4, 9230101H05Rik, Miwi2; piwi-like homolog 4 (... 30.8 1.6
ath:AT4G02410 lectin protein kinase family protein 29.6 3.0
dre:100147850 pgap1, si:dkey-191g15.7; post-GPI attachment to ... 29.6 3.8
hsa:142679 DUSP19, DUSP17, LMWDSP3, MGC138210, SKRP1, TS-DSP1;... 28.9 6.4
ath:AT4G02420 lectin protein kinase, putative 28.5 8.1
> tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8);
K04482 DNA repair protein RAD51
Length=354
Score = 139 bits (349), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 64/86 (74%), Positives = 78/86 (90%), Gaps = 0/86 (0%)
Query 60 LQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQA 119
+Q+GPLKLEHLLAKG T++DL+LLK+ G TVEC+AFAP+K L+A+KG+SEQK KLK+A
Sbjct 31 VQSGPLKLEHLLAKGFTKRDLELLKDAGYQTVECIAFAPVKNLVAVKGLSEQKVEKLKKA 90
Query 120 SKELCSLGFCSAQEYLEARANLIKFT 145
SKELC+LGFCSAQEYLEAR NLI+FT
Sbjct 91 SKELCNLGFCSAQEYLEARENLIRFT 116
> pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein
RAD51
Length=350
Score = 129 bits (323), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 66/107 (61%), Positives = 80/107 (74%), Gaps = 10/107 (9%)
Query 39 SSTVQETRQAPEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAP 98
SST+ E + E L TGPLK+E LLAKG ++DL+LLKEGGL TVECVA+AP
Sbjct 17 SSTIDEIEE----------EQLYTGPLKIEQLLAKGFVKRDLELLKEGGLQTVECVAYAP 66
Query 99 LKALLAIKGISEQKAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
++ L AIKGISEQKA KLK+A KELC+ GFC+A +Y +AR NLIKFT
Sbjct 67 MRTLCAIKGISEQKAEKLKKACKELCNSGFCNAIDYHDARQNLIKFT 113
> cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51
Length=347
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 57/84 (67%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 62 TGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASK 121
GPLKLEHLL GLT++DL++L+E G HT+EC+A+AP KALL++KGISEQK K+K A K
Sbjct 26 NGPLKLEHLLPSGLTKRDLEILRENGYHTIECLAYAPKKALLSVKGISEQKCDKIKSACK 85
Query 122 ELCSLGFCSAQEYLEARANLIKFT 145
EL ++GFCS EYLEAR NLIKFT
Sbjct 86 ELVAMGFCSGTEYLEARTNLIKFT 109
> tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair protein
RAD51
Length=343
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 58/96 (60%), Positives = 70/96 (72%), Gaps = 0/96 (0%)
Query 50 EQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGIS 109
E+ V E P +LE LL+KGL ++DLDLL+E G T+ECVA+AP K LL IKG+S
Sbjct 11 EENSMGVSETGSQNPQRLECLLSKGLLQRDLDLLREAGYSTLECVAYAPQKNLLVIKGLS 70
Query 110 EQKAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
EQK K+K A +ELC LGFCS Q+YLEAR NLIKFT
Sbjct 71 EQKVLKIKAACRELCHLGFCSGQDYLEARGNLIKFT 106
> bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair
protein RAD51
Length=346
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/79 (51%), Positives = 55/79 (69%), Gaps = 0/79 (0%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSL 126
+E LL+KG ++D+D+LK G T++ +A K LL +KG+SEQK AK+K+ KELC
Sbjct 28 VECLLSKGFLQRDIDVLKAAGYVTLDSIAQVASKTLLEVKGLSEQKVAKIKEIVKELCPP 87
Query 127 GFCSAQEYLEARANLIKFT 145
C+A EYLE R NLIKFT
Sbjct 88 DICTAAEYLECRLNLIKFT 106
> ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-dependent
ATPase/ damaged DNA binding / nucleoside-triphosphatase/
nucleotide binding / protein binding / sequence-specific DNA
binding; K04482 DNA repair protein RAD51
Length=342
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/108 (40%), Positives = 61/108 (56%), Gaps = 2/108 (1%)
Query 38 MSSTVQETRQAPEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFA 97
M++ Q Q QQ D E+ Q GP +E L A G+ D+ L++ GL TVE VA+
Sbjct 1 MTTMEQRRNQNAVQQQDD--EETQHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYT 58
Query 98 PLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQEYLEARANLIKFT 145
P K LL IKGIS+ K K+ +A+ +L LGF SA + R +I+ T
Sbjct 59 PRKDLLQIKGISDAKVDKIVEAASKLVPLGFTSASQLHAQRQEIIQIT 106
> dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae); K04482 DNA repair protein
RAD51
Length=338
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 53/83 (63%), Gaps = 0/83 (0%)
Query 63 GPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKE 122
GP + L G++ D+ L++GG HTVE VA+AP K LL IKGISE KA K+ + +
Sbjct 20 GPQPVSRLEQSGISSSDIKKLEDGGFHTVEAVAYAPKKELLNIKGISEAKADKILTEAAK 79
Query 123 LCSLGFCSAQEYLEARANLIKFT 145
+ +GF +A E+ + RA +I+ +
Sbjct 80 MVPMGFTTATEFHQRRAEIIQIS 102
> xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad51;
RAD51 homolog (RecA homolog); K04482 DNA repair protein
RAD51
Length=336
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLK 117
E+ GP + L G+ D+ L+E G HTVE VA+AP K LL IKGISE KA K+
Sbjct 13 EEEHFGPQAISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELLNIKGISEAKAEKIL 72
Query 118 QASKELCSLGFCSAQEYLEARANLIKFT 145
+ +L +GF +A E+ + R+ +I+ +
Sbjct 73 AEAAKLVPMGFTTATEFHQRRSEIIQIS 100
> mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cerevisiae);
K04482 DNA repair protein RAD51
Length=339
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLK 117
E+ GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+
Sbjct 16 EEESFGPQPISRLEQCGINANDVKKLEEAGYHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query 118 QASKELCSLGFCSAQEYLEARANLIKFT 145
+ +L +GF +A E+ + R+ +I+ T
Sbjct 76 TEAAKLVPMGFTTATEFHQRRSEIIQIT 103
> sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51
Length=400
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 49/81 (60%), Gaps = 0/81 (0%)
Query 65 LKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELC 124
+ +E L G+T D+ L+E GLHT E VA+AP K LL IKGISE KA KL + L
Sbjct 81 VPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKDLLEIKGISEAKADKLLNEAARLV 140
Query 125 SLGFCSAQEYLEARANLIKFT 145
+GF +A ++ R+ LI T
Sbjct 141 PMGFVTAADFHMRRSELICLT 161
> xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,
reca, xrad51; RAD51 homolog (RecA homolog); K04482 DNA repair
protein RAD51
Length=336
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 0/87 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLK 117
E+ GP + L G+ D+ L++ G HTVE VA+AP K LL IKGISE KA K+
Sbjct 13 EEENFGPQAITRLEQCGINANDVKKLEDAGFHTVEAVAYAPKKELLNIKGISEAKAEKIL 72
Query 118 QASKELCSLGFCSAQEYLEARANLIKF 144
+ +L +GF +A E+ + R+ +I+
Sbjct 73 AEAAKLVPMGFTTATEFHQRRSEIIQI 99
> hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA;
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae); K04482
DNA repair protein RAD51
Length=340
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLK 117
E+ GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+
Sbjct 16 EEESFGPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKIL 75
Query 118 QASKELCSL 126
S+ + L
Sbjct 76 TESRSVARL 84
> ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1);
ATP binding / DNA binding / DNA-dependent ATPase/ damaged DNA
binding / nucleoside-triphosphatase/ nucleotide binding /
protein binding; K10872 meiotic recombination protein DMC1
Length=344
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSL 126
++ L+A+G+ D+ L+E G+HT + K L IKG+SE K K+ +A++++ +
Sbjct 31 IDKLIAQGINAGDVKKLQEAGIHTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKIVNF 90
Query 127 GFCSAQEYLEARANLIKFT 145
G+ + + L R +++K T
Sbjct 91 GYMTGSDALIKRKSVVKIT 109
> sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination
protein DMC1
Length=334
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 44/84 (52%), Gaps = 0/84 (0%)
Query 55 DVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAA 114
++ D L ++ L G+ DL LK GG++TV V + L IKG+SE K
Sbjct 7 EIDSDTAKNILSVDELQNYGINASDLQKLKSGGIYTVNTVLSTTRRHLCKIKGLSEVKVE 66
Query 115 KLKQASKELCSLGFCSAQEYLEAR 138
K+K+A+ ++ +GF A L+ R
Sbjct 67 KIKEAAGKIIQVGFIPATVQLDIR 90
> dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=342
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSL 126
+E L G+ D+ LK G+ TV+ + +AL IKG+SE K K+K+A+ +L +
Sbjct 26 IELLQKHGINVADIKKLKSVGICTVKGIQMTTRRALCNIKGLSEAKVDKIKEAAGKLLTC 85
Query 127 GFCSAQEYLEARANLIKFT 145
GF +A EY R + T
Sbjct 86 GFQTASEYCIKRKQVFHIT 104
> hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473,
dJ199H16.1; DMC1 dosage suppressor of mck1 homolog, meiosis-specific
homologous recombination (yeast); K10872 meiotic recombination
protein DMC1
Length=340
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 40/72 (55%), Gaps = 0/72 (0%)
Query 74 GLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQE 133
G+ D+ LK G+ T++ + +AL +KG+SE K K+K+A+ +L GF +A E
Sbjct 31 GINVADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKLIEPGFLTAFE 90
Query 134 YLEARANLIKFT 145
Y E R + T
Sbjct 91 YSEKRKMVFHIT 102
> mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=340
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 0/72 (0%)
Query 74 GLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQE 133
G+ D+ LK G+ T++ + +AL +KG+SE K K+K+A+ +L GF +A +
Sbjct 31 GINMADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVEKIKEAANKLIEPGFLTAFQ 90
Query 134 YLEARANLIKFT 145
Y E R + T
Sbjct 91 YSERRKMVFHIT 102
> cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-51); K04482 DNA repair protein
RAD51
Length=395
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 83 LKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLGFCSAQEYLEARANLI 142
LKE G +T E +AF + L +KGIS+QKA K+ + + + +GF + E R+ L+
Sbjct 95 LKEAGYYTYESLAFTTRRELRNVKGISDQKAEKIMKEAMKFVQMGFTTGAEVHVKRSQLV 154
Query 143 KF 144
+
Sbjct 155 QI 156
> cpv:cgd7_1690 meiotic recombination protein DMC1-like protein
; K10872 meiotic recombination protein DMC1
Length=342
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 47/82 (57%), Gaps = 1/82 (1%)
Query 65 LKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELC 124
++++ L + G+ D++ LK GL TV + A K L IKG+SE K K+ +A+++L
Sbjct 25 VEIDKLQSAGINVADINKLKTAGLCTVLSIIQATKKELCNIKGLSEAKVEKIVEAAQKLD 84
Query 125 -SLGFCSAQEYLEARANLIKFT 145
S F S E + R N+++ T
Sbjct 85 QSSSFQSGSEVMSRRQNILRIT 106
> tgo:TGME49_016400 meiotic recombination protein DMC1-like protein,
putative (EC:2.7.11.1); K10872 meiotic recombination
protein DMC1
Length=349
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 67 LEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKEL--C 124
++ L A G+ D++ LK+ G TV + K L +KGISE K K+ +A+ +L C
Sbjct 34 IDKLQAAGINAADINKLKQAGYCTVLSIVQTTKKELCLVKGISEAKVEKIVEAAAKLGMC 93
Query 125 SLGFCSAQEYLEARANLIKFT 145
+ F + E ++ R +IK T
Sbjct 94 N-AFITGVELVQKRGRVIKIT 113
> pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;
K10872 meiotic recombination protein DMC1
Length=347
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 66 KLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAK-LKQASK-EL 123
++E L G+ D++ LK G T+ + K L +KGISE K K L+ ASK E
Sbjct 31 EIEKLQDLGINAADINKLKGSGYCTILSLIQTTKKELCNVKGISEAKVDKILEVASKIEN 90
Query 124 CSLGFCSAQEYLEARANLIKFT 145
CS F +A E ++ R+ ++K T
Sbjct 91 CS-SFITANELVQKRSKVLKIT 111
> eco:b3169 nusA, ECK3158, JW3138; transcription termination/antitermination
L factor; K02600 N utilization substance protein
A
Length=495
Score = 33.5 bits (75), Expect = 0.25, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 82 LLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASK 121
+L E G T+E +A+ P+K LL I+G+ E L++ +K
Sbjct 372 VLVEEGFSTLEELAYVPMKELLEIEGLDEPTVEALRERAK 411
Score = 28.5 bits (62), Expect = 6.6, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 48/102 (47%), Gaps = 12/102 (11%)
Query 28 PTAHGLDFRPMSSTVQETRQAPEQQLQD--VGEDLQTGPLKLEHLLAKGLTRKDLDLLKE 85
PT L R ++ + QA E+ L D +DL L LE G+ R L
Sbjct 401 PTVEALRERAKNA-LATIAQAQEESLGDNKPADDL----LNLE-----GVDRDLAFKLAA 450
Query 86 GGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLG 127
G+ T+E +A + L I+G++++KA L A++ +C G
Sbjct 451 RGVCTLEDLAEQGIDDLADIEGLTDEKAGALIMAARNICWFG 492
> mmu:330890 Piwil4, 9230101H05Rik, Miwi2; piwi-like homolog 4
(Drosophila); K02156 aubergine
Length=878
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 34/68 (50%), Gaps = 11/68 (16%)
Query 5 LLQSSLRRHRRGGG-------LPSLCFYLWPTAHGL-DFRPMSSTVQETRQAP---EQQL 53
+L S L+R R +P LCF ++ DFR M + +ETR +P +QQL
Sbjct 397 VLVSLLKRKRNDNSEPQMVHLMPELCFLTGLSSQATSDFRLMKAVAEETRLSPVGRQQQL 456
Query 54 QDVGEDLQ 61
+ +D+Q
Sbjct 457 ARLVDDIQ 464
> ath:AT4G02410 lectin protein kinase family protein
Length=674
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 27/120 (22%), Positives = 48/120 (40%), Gaps = 1/120 (0%)
Query 8 SSLRRHRRGGGLPSLCFYLWPTAHGLDFRPMSSTVQETRQAPEQQLQDVGEDLQTGPLKL 67
+SL+R + +P L P + + + R+ ++ +D + L+
Sbjct 287 TSLQRFYKNR-MPLFSLLLIPVLFVVSLIFLVRFIVRRRRKFAEEFEDWETEFGKNRLRF 345
Query 68 EHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAKLKQASKELCSLG 127
+ L KD DLL GG V K +A+K +S + LK+ E+ S+G
Sbjct 346 KDLYYATKGFKDKDLLGSGGFGRVYRGVMPTTKKEIAVKRVSNESRQGLKEFVAEIVSIG 405
> dre:100147850 pgap1, si:dkey-191g15.7; post-GPI attachment to
proteins 1; K05294 glycosylphosphatidylinositol deacylase
[EC:3.-.-.-]
Length=1031
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 58 EDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKALLAIKGISEQKAAK 115
E+ T +++ HLL+ GLT D+ + G LHT++ F + I +S K K
Sbjct 550 EESLTVSVQVPHLLSFGLTVTDVSINSSGLLHTLQLQDFHQVYQAFRITIMSHCKTNK 607
> hsa:142679 DUSP19, DUSP17, LMWDSP3, MGC138210, SKRP1, TS-DSP1;
dual specificity phosphatase 19 (EC:3.1.3.16 3.1.3.48); K14165
dual specificity phosphatase [EC:3.1.3.16 3.1.3.48]
Length=166
Score = 28.9 bits (63), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 53 LQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEG--GLHTVECVAFAPLKALLAI 105
+QD+ DLQ G +K LL DLD LK+ G+ V C A A + I
Sbjct 57 VQDLSSDLQVGVIKPWLLLGSQDAAHDLDTLKKNKDGVVLVHCNAGVSRAAAIVI 111
> ath:AT4G02420 lectin protein kinase, putative
Length=669
Score = 28.5 bits (62), Expect = 8.1, Method: Composition-based stats.
Identities = 19/86 (22%), Positives = 39/86 (45%), Gaps = 0/86 (0%)
Query 42 VQETRQAPEQQLQDVGEDLQTGPLKLEHLLAKGLTRKDLDLLKEGGLHTVECVAFAPLKA 101
+ + R+ ++++D + L+ + L KD ++L GG +V K
Sbjct 315 IMKRRRKFAEEVEDWETEFGKNRLRFKDLYYATKGFKDKNILGSGGFGSVYKGIMPKTKK 374
Query 102 LLAIKGISEQKAAKLKQASKELCSLG 127
+A+K +S + LK+ E+ S+G
Sbjct 375 EIAVKRVSNESRQGLKEFVAEIVSIG 400
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2807590228
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40