bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0604_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
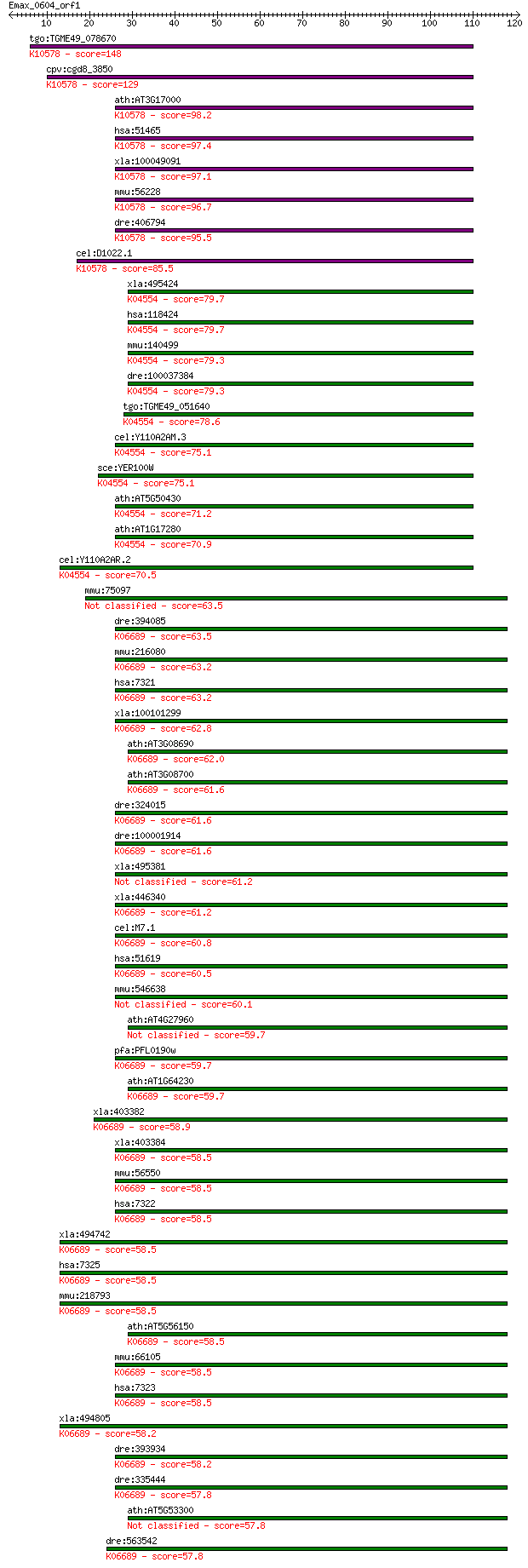
tgo:TGME49_078670 ubiquitin-conjugating enzyme E2, putative (E... 148 5e-36
cpv:cgd8_3850 Ubc6p like ubiquiting conjugating enzyme E2, pos... 129 2e-30
ath:AT3G17000 UBC32; UBC32 (ubiquitin-conjugating enzyme 32); ... 98.2 6e-21
hsa:51465 UBE2J1, HSPC153, HSPC205, HSU93243, MGC12555, NCUBE-... 97.4 1e-20
xla:100049091 ube2j1; ubiquitin-conjugating enzyme E2, J1 (UBC... 97.1 1e-20
mmu:56228 Ube2j1, 0710008M05Rik, 1110030I22Rik, NCUBE-1, Ncube... 96.7 2e-20
dre:406794 zgc:63554 (EC:6.3.2.19); K10578 ubiquitin-conjugati... 95.5 4e-20
cel:D1022.1 ubc-6; UBiquitin Conjugating enzyme family member ... 85.5 3e-17
xla:495424 ube2j2; ubiquitin-conjugating enzyme E2, J2 (UBC6 h... 79.7 2e-15
hsa:118424 UBE2J2, NCUBE-2, NCUBE2, PRO2121; ubiquitin-conjuga... 79.7 2e-15
mmu:140499 Ube2j2, 1200007B18Rik, 2400008G19Rik, 5730472G04Rik... 79.3 3e-15
dre:100037384 ube2j2, zgc:162164; ubiquitin-conjugating enzyme... 79.3 3e-15
tgo:TGME49_051640 ubiquitin-conjugating enzyme E2, putative (E... 78.6 4e-15
cel:Y110A2AM.3 ubc-26; UBiquitin Conjugating enzyme family mem... 75.1 5e-14
sce:YER100W UBC6, DOA2; Ubc6p (EC:6.3.2.19); K04554 ubiquitin-... 75.1 6e-14
ath:AT5G50430 UBC33; UBC33 (ubiquitin-conjugating enzyme 33); ... 71.2 7e-13
ath:AT1G17280 UBC34; UBC34 (ubiquitin-conjugating enzyme 34); ... 70.9 1e-12
cel:Y110A2AR.2 ubc-15; UBiquitin Conjugating enzyme family mem... 70.5 1e-12
mmu:75097 4930524E20Rik; RIKEN cDNA 4930524E20 gene 63.5 2e-10
dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689 ub... 63.5 2e-10
mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzy... 63.2 2e-10
hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiqui... 63.2 2e-10
xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating ... 62.8 2e-10
ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11); ... 62.0 5e-10
ath:AT3G08700 UBC12; UBC12 (ubiquitin-conjugating enzyme 12); ... 61.6 5e-10
dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugatin... 61.6 5e-10
dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjuga... 61.6 5e-10
xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiq... 61.2 8e-10
xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D ... 61.2 8e-10
cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquit... 60.8 1e-09
hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzy... 60.5 1e-09
mmu:546638 Gm5959, EG546638; predicted gene 5959 60.1
ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubi... 59.7 2e-09
pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3... 59.7 2e-09
ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative; K... 59.7 2e-09
xla:403382 ube2e3-b, ubc15, ubch9, ubcm2, xubc15; ubiquitin-co... 58.9 4e-09
xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugat... 58.5 5e-09
mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjug... 58.5 5e-09
hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiqu... 58.5 5e-09
xla:494742 ube2e2, ubch8; ubiquitin-conjugating enzyme E2E 2 (... 58.5 5e-09
hsa:7325 UBE2E2, FLJ25157, UBCH8; ubiquitin-conjugating enzyme... 58.5 5e-09
mmu:218793 Ube2e2, BC016265, MGC28917; ubiquitin-conjugating e... 58.5 5e-09
ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30); ... 58.5 5e-09
mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiq... 58.5 5e-09
hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;... 58.5 5e-09
xla:494805 hypothetical LOC494805; K06689 ubiquitin-conjugatin... 58.2 6e-09
dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19... 58.2 6e-09
dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugatin... 57.8 8e-09
ath:AT5G53300 UBC10; UBC10 (ubiquitin-conjugating enzyme 10); ... 57.8 8e-09
dre:563542 MGC171646; zgc:171646 (EC:6.3.2.-); K06689 ubiquiti... 57.8 9e-09
> tgo:TGME49_078670 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=201
Score = 148 bits (373), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 70/107 (65%), Positives = 83/107 (77%), Gaps = 3/107 (2%)
Query 6 MEVAHDTGRAGAAQ---HASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTL 62
M+ + GR G + +TAQ +ARILRE R+IQR SPHW A PL +EEP EWHFTL
Sbjct 1 MQGVSNPGRTGTNANLGYTATAQCIARILREFREIQRTPSPHWCANPLQIEEPYEWHFTL 60
Query 63 RGPPDSPFEGGLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKKVRL 109
RGP DS FEGGLYHGRIVLPKNYPFAPP+L++LT+NGRF++ KKV L
Sbjct 61 RGPQDSHFEGGLYHGRIVLPKNYPFAPPNLVMLTQNGRFEVGKKVCL 107
> cpv:cgd8_3850 Ubc6p like ubiquiting conjugating enzyme E2, possible
transmembrane domain at C ; K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=368
Score = 129 bits (324), Expect = 2e-30, Method: Composition-based stats.
Identities = 57/101 (56%), Positives = 77/101 (76%), Gaps = 1/101 (0%)
Query 10 HDTGRAGAAQHASTA-QSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDS 68
H TG G T+ Q L+RILRE+R+IQ+ S +W A P++++EP EWHFT++GP +
Sbjct 34 HQTGARGVGSSGITSVQCLSRILREYREIQKEPSSYWCAFPINMDEPYEWHFTIKGPAGT 93
Query 69 PFEGGLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKKVRL 109
FEGG+YHGRI+LP +YPF+PPSLM+LT NGRF++ KKV L
Sbjct 94 EFEGGMYHGRIILPHSYPFSPPSLMMLTGNGRFEVGKKVCL 134
> ath:AT3G17000 UBC32; UBC32 (ubiquitin-conjugating enzyme 32);
ubiquitin-protein ligase; K10578 ubiquitin-conjugating enzyme
E2 J1 [EC:6.3.2.19]
Length=309
Score = 98.2 bits (243), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 59/84 (70%), Gaps = 1/84 (1%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
++ RIL+E +++Q N S + + PL E EW F +RGP D+ FEGG+YHGRI LP +Y
Sbjct 12 AVKRILQEVKEMQANPSDDFMSLPLE-ENIFEWQFAIRGPGDTEFEGGIYHGRIQLPADY 70
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
PF PPS MLLT NGRF+ N K+ L
Sbjct 71 PFKPPSFMLLTPNGRFETNTKICL 94
> hsa:51465 UBE2J1, HSPC153, HSPC205, HSU93243, MGC12555, NCUBE-1,
NCUBE1, Ubc6p; ubiquitin-conjugating enzyme E2, J1 (UBC6
homolog, yeast) (EC:6.3.2.19); K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=318
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 43/84 (51%), Positives = 63/84 (75%), Gaps = 2/84 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
++ R+++E ++ ++ + H+ A PL + EWHFT+RGPPDS F+GG+YHGRIVLP Y
Sbjct 11 AVKRLMKEAAEL-KDPTDHYHAQPLE-DNLFEWHFTVRGPPDSDFDGGVYHGRIVLPPEY 68
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P PPS++LLT NGRF++ KK+ L
Sbjct 69 PMKPPSIILLTANGRFEVGKKICL 92
> xla:100049091 ube2j1; ubiquitin-conjugating enzyme E2, J1 (UBC6
homolog); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=302
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 44/84 (52%), Positives = 63/84 (75%), Gaps = 2/84 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
++ R+++E ++ R+ + H+ A PL + EWHFT+RGPPDS F+GG+YHGRIVLP Y
Sbjct 11 AVKRLMKEAAEL-RDPTDHYHAQPLE-DNLFEWHFTVRGPPDSDFDGGVYHGRIVLPPEY 68
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P PPS++LLT NGRF++ KK+ L
Sbjct 69 PMKPPSIILLTANGRFEVGKKICL 92
> mmu:56228 Ube2j1, 0710008M05Rik, 1110030I22Rik, NCUBE-1, Ncube,
Ncube1, Ubc6p; ubiquitin-conjugating enzyme E2, J1 (EC:6.3.2.19);
K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=318
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 43/84 (51%), Positives = 63/84 (75%), Gaps = 2/84 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
++ R+++E ++ ++ + H+ A PL + EWHFT+RGPPDS F+GG+YHGRIVLP Y
Sbjct 11 AVKRLMKEAAEL-KDPTDHYHAQPLE-DNLFEWHFTVRGPPDSDFDGGVYHGRIVLPPEY 68
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P PPS++LLT NGRF++ KK+ L
Sbjct 69 PMKPPSIILLTANGRFEVGKKICL 92
> dre:406794 zgc:63554 (EC:6.3.2.19); K10578 ubiquitin-conjugating
enzyme E2 J1 [EC:6.3.2.19]
Length=314
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 43/84 (51%), Positives = 63/84 (75%), Gaps = 2/84 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
++ R+++E ++ R+ + H+ A PL + EWHF++RGPPDS F+GG+YHGRIVLP Y
Sbjct 11 AVKRLMKEAAEL-RDPTEHYHAQPLE-DNLFEWHFSVRGPPDSDFDGGVYHGRIVLPPEY 68
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P PPS++LLT NGRF++ KK+ L
Sbjct 69 PMKPPSIILLTPNGRFEVGKKICL 92
> cel:D1022.1 ubc-6; UBiquitin Conjugating enzyme family member
(ubc-6); K10578 ubiquitin-conjugating enzyme E2 J1 [EC:6.3.2.19]
Length=314
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 62/93 (66%), Gaps = 2/93 (2%)
Query 17 AAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYH 76
+ Q+ + + R+++E ++ R + + A P+ + EWHFT+RG + FEGG+YH
Sbjct 2 SEQYNTKNAGVRRLMKEAMEL-RQPTEMYHAQPME-DNLFEWHFTIRGTLGTDFEGGIYH 59
Query 77 GRIVLPKNYPFAPPSLMLLTRNGRFDINKKVRL 109
GRI+ P +YP PP+L+LLT NGRF++NKKV L
Sbjct 60 GRIIFPADYPMKPPNLILLTPNGRFELNKKVCL 92
> xla:495424 ube2j2; ubiquitin-conjugating enzyme E2, J2 (UBC6
homolog); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
R+ +++ I+++ P+ A PL EWH+ +RGP +P+EGG YHG++V P+ +PF
Sbjct 16 RLKQDYLRIKKDPVPYICAEPLP-SNILEWHYVVRGPEMTPYEGGYYHGKLVFPREFPFK 74
Query 89 PPSLMLLTRNGRFDINKKVRL 109
PPS+ ++T NGRF N ++ L
Sbjct 75 PPSIYMITPNGRFKCNTRLCL 95
> hsa:118424 UBE2J2, NCUBE-2, NCUBE2, PRO2121; ubiquitin-conjugating
enzyme E2, J2 (UBC6 homolog, yeast) (EC:6.3.2.19); K04554
ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
R+ +++ I+++ P+ A PL EWH+ +RGP +P+EGG YHG+++ P+ +PF
Sbjct 16 RLKQDYLRIKKDPVPYICAEPLP-SNILEWHYVVRGPEMTPYEGGYYHGKLIFPREFPFK 74
Query 89 PPSLMLLTRNGRFDINKKVRL 109
PPS+ ++T NGRF N ++ L
Sbjct 75 PPSIYMITPNGRFKCNTRLCL 95
> mmu:140499 Ube2j2, 1200007B18Rik, 2400008G19Rik, 5730472G04Rik,
AL022923, NCUBE-2, Ubc6, Ubc6p; ubiquitin-conjugating enzyme
E2, J2 homolog (yeast) (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=271
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
R+ +++ I+++ P+ A PL EWH+ +RGP +P+EGG YHG+++ P+ +PF
Sbjct 28 RLKQDYLRIKKDPVPYICAEPLP-SNILEWHYVVRGPEMTPYEGGYYHGKLIFPREFPFK 86
Query 89 PPSLMLLTRNGRFDINKKVRL 109
PPS+ ++T NGRF N ++ L
Sbjct 87 PPSIYMITPNGRFKCNTRLCL 107
> dre:100037384 ube2j2, zgc:162164; ubiquitin-conjugating enzyme
E2, J2 (UBC6 homolog, yeast) (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=259
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
R+ +++ I+++ P+ A PL EWH+ +RGP +P+EGG YHG+++ P+ +PF
Sbjct 16 RLKQDYLRIKKDPVPYICAEPLP-SNILEWHYLVRGPEKTPYEGGYYHGKLIFPREFPFK 74
Query 89 PPSLMLLTRNGRFDINKKVRL 109
PPS+ ++T NGRF N ++ L
Sbjct 75 PPSIYMITPNGRFKCNTRLCL 95
> tgo:TGME49_051640 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=275
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 53/84 (63%), Gaps = 3/84 (3%)
Query 28 ARILREHRDIQRNAS-PHWTAAPLSLEEPREWHFTLRG-PPDSPFEGGLYHGRIVLPKNY 85
R+ RE +QR PH P + + WHF L P DSP+ GG+YHG++V P NY
Sbjct 16 GRLAREFSLLQRQGGVPHAQLQP-DMNDTLTWHFVLHDLPADSPYHGGVYHGKLVFPPNY 74
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
PF PPS+ +LT +GRF++NK++ +
Sbjct 75 PFEPPSIFMLTPSGRFEVNKRICM 98
> cel:Y110A2AM.3 ubc-26; UBiquitin Conjugating enzyme family member
(ubc-26); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=203
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L R+ ++++ + + P AAPL EW + + G P +P+EGG+Y G+++ PK++
Sbjct 9 ALQRLKKDYQRLLKEPVPFMKAAPLE-TNILEWRYIIIGAPKTPYEGGIYMGKLLFPKDF 67
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
PF PP++++LT NGRF N ++ L
Sbjct 68 PFKPPAILMLTPNGRFQTNTRLCL 91
> sce:YER100W UBC6, DOA2; Ubc6p (EC:6.3.2.19); K04554 ubiquitin-conjugating
enzyme E2 J2 [EC:6.3.2.19]
Length=250
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 56/88 (63%), Gaps = 1/88 (1%)
Query 22 STAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVL 81
+T Q+ R+ +E++ + N P+ A P + + EWH+ + GP D+P++GG YHG +
Sbjct 2 ATKQAHKRLTKEYKLMVENPPPYILARP-NEDNILEWHYIITGPADTPYKGGQYHGTLTF 60
Query 82 PKNYPFAPPSLMLLTRNGRFDINKKVRL 109
P +YP+ PP++ ++T NGRF N ++ L
Sbjct 61 PSDYPYKPPAIRMITPNGRFKPNTRLCL 88
> ath:AT5G50430 UBC33; UBC33 (ubiquitin-conjugating enzyme 33);
ubiquitin-protein ligase; K04554 ubiquitin-conjugating enzyme
E2 J2 [EC:6.3.2.19]
Length=243
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+ R+ +E+R + + H A P S + EWH+ L G +PF GG Y+G+I P Y
Sbjct 6 CIKRLQKEYRALCKEPVSHVVARP-SPNDILEWHYVLEGSEGTPFAGGFYYGKIKFPPEY 64
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P+ PP + + T NGRF KK+ L
Sbjct 65 PYKPPGITMTTPNGRFVTQKKICL 88
> ath:AT1G17280 UBC34; UBC34 (ubiquitin-conjugating enzyme 34);
ubiquitin-protein ligase; K04554 ubiquitin-conjugating enzyme
E2 J2 [EC:6.3.2.19]
Length=237
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+ R+ +E+R + + H A P S + EWH+ L G +PF GG Y+G+I P Y
Sbjct 6 CIKRLQKEYRALCKEPVSHVVARP-SPNDILEWHYVLEGSEGTPFAGGFYYGKIKFPPEY 64
Query 86 PFAPPSLMLLTRNGRFDINKKVRL 109
P+ PP + + T NGRF KK+ L
Sbjct 65 PYKPPGITMTTPNGRFMTQKKICL 88
> cel:Y110A2AR.2 ubc-15; UBiquitin Conjugating enzyme family member
(ubc-15); K04554 ubiquitin-conjugating enzyme E2 J2 [EC:6.3.2.19]
Length=218
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 60/97 (61%), Gaps = 1/97 (1%)
Query 13 GRAGAAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEG 72
G + A +++ ++ R+ +++ + ++ A P + + EWH+ LRG PD+PF G
Sbjct 7 GVSVPAAGTASSSAVRRLQKDYAKLMQDPVDGIKALP-NEDNILEWHYCLRGSPDTPFYG 65
Query 73 GLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKKVRL 109
G Y G+++ +N+P++PP++ ++T NGRF N ++ L
Sbjct 66 GYYWGKVIFKENFPWSPPAITMITPNGRFQTNTRLCL 102
> mmu:75097 4930524E20Rik; RIKEN cDNA 4930524E20 gene
Length=155
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 54/100 (54%), Gaps = 2/100 (2%)
Query 19 QHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGR 78
Q A +L RI +E I ++ H +A P++ E W T+ GP DSP++GG++
Sbjct 3 QSTLGAMALKRIQKELVAISQDPPAHCSAGPVA-ENMFHWQATIMGPEDSPYQGGVFFLS 61
Query 79 IVLPKNYPFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
+ P NYPF PP + +TR +I+K + L L+S W
Sbjct 62 VHFPNNYPFKPPKVTFITRVYHPNISKNGSICLDILNSMW 101
> dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E +D+QR+ +A PL E+ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELQDLQRDPPSQCSAGPLG-EDLFHWQATIMGPGDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzyme
E2D 1, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ H +A P+ ++ W T+ GPPDS ++GG++ + P +Y
Sbjct 2 ALKRIQKELSDLQRDPPAHCSAGPVG-DDLFHWQATIMGPPDSAYQGGVFFLTVHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKIAFTTKIYHPNINSNGSICLDILRSQW 93
> hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ H +A P+ ++ W T+ GPPDS ++GG++ + P +Y
Sbjct 2 ALKRIQKELSDLQRDPPAHCSAGPVG-DDLFHWQATIMGPPDSAYQGGVFFLTVHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKIAFTTKIYHPNINSNGSICLDILRSQW 93
> xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ E+ W T+ GP DSPF+GG++ I P +Y
Sbjct 2 ALKRIQKELMDLQRDPPAQCSAGPVG-EDLFHWQATIMGPNDSPFQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 52/90 (57%), Gaps = 2/90 (2%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RIL+E +D+Q++ + +A P++ E+ W T+ GPP+SP+ GG++ I P +YPF
Sbjct 5 RILKELKDLQKDPPSNCSAGPVA-EDMFHWQATIMGPPESPYAGGVFLVSIHFPPDYPFK 63
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ +IN + L L +W
Sbjct 64 PPKVSFKTKVYHPNINSNGSICLDILKEQW 93
> ath:AT3G08700 UBC12; UBC12 (ubiquitin-conjugating enzyme 12);
small conjugating protein ligase/ ubiquitin-protein ligase;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=149
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RI RE RD+QR+ + +A P++ E+ W T+ GP DSP+ GG++ I +YPF
Sbjct 5 RISRELRDMQRHPPANCSAGPVAEEDIFHWQATIMGPHDSPYSGGVFTVSIDFSSDYPFK 64
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ +I+ K + L L +W
Sbjct 65 PPKVNFKTKVYHPNIDSKGSICLDILKEQW 94
> dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E +D+QR+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELQDLQRDPPAQCSAGPVG-DDLFHWQATIMGPSDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ E+ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELTDLQRDPPAQCSAGPVG-EDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog)
Length=147
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELNDLQRDPPAQCSAGPVG-DDLFHWQATIMGPTDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D
2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELNDLQRDPPAQCSAGPVG-DDLFHWQATIMGPTDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E +D+ R+ +A P+ ++ W T+ GPP+SP++GG++ I P +Y
Sbjct 2 ALKRIQKELQDLGRDPPAQCSAGPVG-DDLFHWQATIMGPPESPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzyme
E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIQKELTDLQRDPPAQCSAGPVG-DDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + T+ +IN + L L S+W
Sbjct 61 PFKPPKVAFTTKIYHPNINSNGSICLDILRSQW 93
> mmu:546638 Gm5959, EG546638; predicted gene 5959
Length=179
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+QR+ +A P+ ++ W T+ GP DSP +GG++ I P +Y
Sbjct 2 ALKRIQKEPTDLQRDLPAQCSAGPVG-DDLFHWQATILGPNDSPCQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPEVAFTTRIYHPNINSNGSIYLDTLRSQW 93
> ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubiquitin-protein
ligase
Length=178
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RIL+E +D+Q++ +A P++ E+ W T+ GP DSP+ GG++ I P +YPF
Sbjct 35 RILKELKDLQKDPPTSCSAGPVA-EDMFHWQATIMGPSDSPYSGGVFLVTIHFPPDYPFK 93
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ +IN + L L +W
Sbjct 94 PPKVAFRTKVFHPNINSNGSICLDILKEQW 123
> pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E +D+ ++ + +A P+ ++ W T+ GP DSP+E G+Y I P +Y
Sbjct 2 ALKRITKELQDLNKDPPTNCSAGPIG-DDLFFWQATIMGPGDSPYENGVYFLNIKFPPDY 60
Query 86 PFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
PF PP ++ T+ +IN L L +W
Sbjct 61 PFKPPKIIFTTKIYHPNINTAGAICLDILKDQW 93
> ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RIL+E +D+Q++ +A P++ E+ W T+ GP DSP+ GG++ I P +YPF
Sbjct 5 RILKELKDLQKDPPTSCSAGPVA-EDMFHWQATIMGPSDSPYSGGVFLVTIHFPPDYPFK 63
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ ++N + L L +W
Sbjct 64 PPKVAFRTKVFHPNVNSNGSICLDILKEQW 93
> xla:403382 ube2e3-b, ubc15, ubch9, ubcm2, xubc15; ubiquitin-conjugating
enzyme E2E 3 (UBC4/5 homolog) (EC:6.3.2.19); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=259
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 48/98 (48%), Gaps = 2/98 (2%)
Query 21 ASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIV 80
A + S RI +E DI + P+ +A P + EW T+ GPP S +EGG++ I
Sbjct 109 AKLSTSAKRIQKELADITLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEGGVFFLDIT 167
Query 81 LPKNYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
+YPF PP + TR +IN + V L L W
Sbjct 168 FSSDYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 205
> xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjugating
enzyme E2D 2 (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIHKELNDLARDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> xla:494742 ube2e2, ubch8; ubiquitin-conjugating enzyme E2E 2
(UBC4/5 homolog); K06689 ubiquitin-conjugating enzyme E2 D/E
[EC:6.3.2.19]
Length=201
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 51/106 (48%), Gaps = 2/106 (1%)
Query 13 GRAGAAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEG 72
G+ + A + S RI +E +I + P+ +A P + EW T+ GPP S +EG
Sbjct 43 GKISSKTAAKLSTSAKRIQKELAEITLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEG 101
Query 73 GLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
G++ I +YPF PP + TR +IN + V L L W
Sbjct 102 GVFFLDITFSPDYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 147
> hsa:7325 UBE2E2, FLJ25157, UBCH8; ubiquitin-conjugating enzyme
E2E 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 51/106 (48%), Gaps = 2/106 (1%)
Query 13 GRAGAAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEG 72
G+ + A + S RI +E +I + P+ +A P + EW T+ GPP S +EG
Sbjct 43 GKISSKTAAKLSTSAKRIQKELAEITLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEG 101
Query 73 GLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
G++ I +YPF PP + TR +IN + V L L W
Sbjct 102 GVFFLDITFSPDYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 147
> mmu:218793 Ube2e2, BC016265, MGC28917; ubiquitin-conjugating
enzyme E2E 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 51/106 (48%), Gaps = 2/106 (1%)
Query 13 GRAGAAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEG 72
G+ + A + S RI +E +I + P+ +A P + EW T+ GPP S +EG
Sbjct 43 GKISSKTAAKLSTSAKRIQKELAEITLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEG 101
Query 73 GLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
G++ I +YPF PP + TR +IN + V L L W
Sbjct 102 GVFFLDITFSPDYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 147
> ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 48/90 (53%), Gaps = 2/90 (2%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RI +E RD+QR+ +A P ++ +W T+ GP DSPF GG++ I P +YPF
Sbjct 5 RINKELRDLQRDPPVSCSAGPTG-DDMFQWQATIMGPADSPFAGGVFLVTIHFPPDYPFK 63
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ +IN + L L +W
Sbjct 64 PPKVAFRTKVYHPNINSNGSICLDILKEQW 93
> mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRINKELSDLARDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E
[EC:6.3.2.19]
Length=147
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRINKELSDLARDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> xla:494805 hypothetical LOC494805; K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=201
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 51/106 (48%), Gaps = 2/106 (1%)
Query 13 GRAGAAQHASTAQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEG 72
G+ + A + S RI +E +I + P+ +A P + EW T+ GPP S +EG
Sbjct 43 GKISSKTAAKLSTSAKRIQKELAEITLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEG 101
Query 73 GLYHGRIVLPKNYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
G++ I +YPF PP + TR +IN + V L L W
Sbjct 102 GVFFLDIAFSPDYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 147
> dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIHKELHDLGRDPPAQCSAGPVG-DDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query 26 SLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNY 85
+L RI +E D+ R+ +A P+ ++ W T+ GP DSP++GG++ I P +Y
Sbjct 2 ALKRIHKELTDLGRDPPAQCSAGPVG-DDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDY 60
Query 86 PFAPPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PF PP + TR +IN + L L S+W
Sbjct 61 PFKPPKVAFTTRIYHPNINSNGSICLDILRSQW 93
> ath:AT5G53300 UBC10; UBC10 (ubiquitin-conjugating enzyme 10);
ubiquitin-protein ligase
Length=109
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 29 RILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPKNYPFA 88
RIL+E +D+Q++ +A P++ E+ W T+ GP +SP+ GG++ I P +YPF
Sbjct 5 RILKELKDLQKDPPTSCSAGPVA-EDMFHWQATIMGPSESPYAGGVFLVTIHFPPDYPFK 63
Query 89 PPSLMLLTRNGRFDINKKVRL-LSALSSRW 117
PP + T+ +IN + L L +W
Sbjct 64 PPKVAFRTKVFHPNINSNGSICLDILKEQW 93
> dre:563542 MGC171646; zgc:171646 (EC:6.3.2.-); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=195
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 47/95 (49%), Gaps = 2/95 (2%)
Query 24 AQSLARILREHRDIQRNASPHWTAAPLSLEEPREWHFTLRGPPDSPFEGGLYHGRIVLPK 83
+ S RI +E DI + P+ +A P + EW T+ GPP S +EGG++ I
Sbjct 48 STSAKRIQKELADIMLDPPPNCSAGPKG-DNIYEWRSTILGPPGSVYEGGVFFLDIAFTP 106
Query 84 NYPFAPPSLMLLTRNGRFDINKK-VRLLSALSSRW 117
+YPF PP + TR +IN + V L L W
Sbjct 107 DYPFKPPKVTFRTRIYHCNINSQGVICLDILKDNW 141
Lambda K H
0.321 0.135 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40