bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
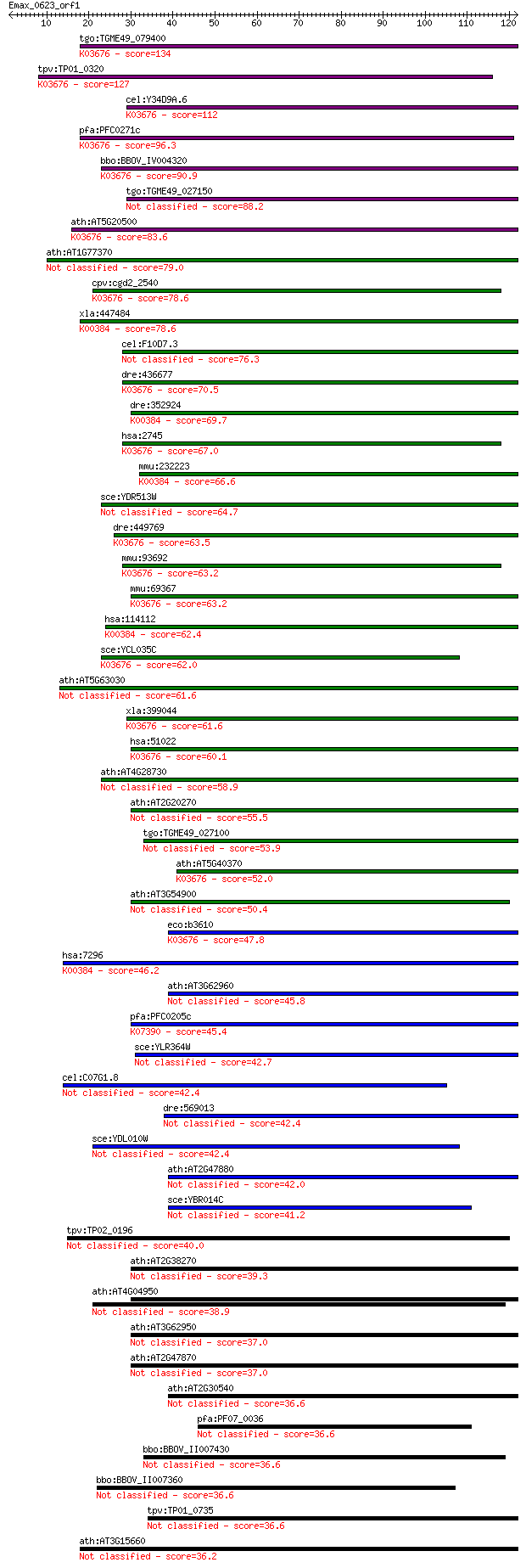
Query= Emax_0623_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_079400 glutaredoxin, putative ; K03676 glutaredoxin 3 134 9e-32
tpv:TP01_0320 glutaredoxin-like protein; K03676 glutaredoxin 3 127 7e-30
cel:Y34D9A.6 glrx-10; GLutaRedoXin family member (glrx-10); K0... 112 3e-25
pfa:PFC0271c GRX1, GRX-1; glutaredoxin 1; K03676 glutaredoxin 3 96.3
bbo:BBOV_IV004320 23.m06161; glutaredoxin-like protein; K03676... 90.9 9e-19
tgo:TGME49_027150 glutaredoxin, putative 88.2 5e-18
ath:AT5G20500 glutaredoxin, putative; K03676 glutaredoxin 3 83.6
ath:AT1G77370 glutaredoxin, putative 79.0 3e-15
cpv:cgd2_2540 glutaredoxin related protein ; K03676 glutaredox... 78.6 4e-15
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 78.6 5e-15
cel:F10D7.3 hypothetical protein 76.3 2e-14
dre:436677 glrx2, zgc:92698; glutaredoxin 2; K03676 glutaredox... 70.5 1e-12
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 69.7 2e-12
hsa:2745 GLRX, GRX, GRX1, MGC117407; glutaredoxin (thioltransf... 67.0 2e-11
mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase 3... 66.6 2e-11
sce:YDR513W GRX2, TTR1; Cytoplasmic glutaredoxin, thioltransfe... 64.7 7e-11
dre:449769 glrx, MGC113837, grx, wu:fc38f02, zgc:103707; gluta... 63.5 1e-10
mmu:93692 Glrx, C86710, D13Wsu156e, Glrx1, Grx1, TTase; glutar... 63.2 2e-10
mmu:69367 Glrx2, 1700010P22Rik, AI645710, Grx2; glutaredoxin 2... 63.2 2e-10
hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3 (E... 62.4 4e-10
sce:YCL035C GRX1; Grx1p; K03676 glutaredoxin 3 62.0
ath:AT5G63030 glutaredoxin, putative 61.6 5e-10
xla:399044 glrx, MGC68461, grx, grx1; glutaredoxin (thioltrans... 61.6 6e-10
hsa:51022 GLRX2, GRX2, bA101E13.1; glutaredoxin 2; K03676 glut... 60.1 2e-09
ath:AT4G28730 glutaredoxin family protein 58.9 4e-09
ath:AT2G20270 glutaredoxin family protein 55.5 4e-08
tgo:TGME49_027100 glutaredoxin, putative 53.9 1e-07
ath:AT5G40370 glutaredoxin, putative; K03676 glutaredoxin 3 52.0
ath:AT3G54900 CXIP1; CXIP1 (CAX INTERACTING PROTEIN 1); antipo... 50.4 1e-06
eco:b3610 grxC, ECK3600, JW3585, yibM; glutaredoxin 3; K03676 ... 47.8 8e-06
hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thior... 46.2 3e-05
ath:AT3G62960 glutaredoxin family protein 45.8 3e-05
pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K0739... 45.4 4e-05
sce:YLR364W GRX8; Glutaredoxin that employs a dithiol mechanis... 42.7 3e-04
cel:C07G1.8 glrx-22; GLutaRedoXin family member (glrx-22) 42.4 3e-04
dre:569013 MGC152951; zgc:152951 42.4 4e-04
sce:YDL010W GRX6; Cis-golgi localized monothiol glutaredoxin t... 42.4 4e-04
ath:AT2G47880 glutaredoxin family protein 42.0 4e-04
sce:YBR014C GRX7; Cis-golgi localized monothiol glutaredoxin; ... 41.2 8e-04
tpv:TP02_0196 hypothetical protein 40.0 0.002
ath:AT2G38270 CXIP2; CXIP2 (CAX-INTERACTING PROTEIN 2); electr... 39.3 0.003
ath:AT4G04950 thioredoxin family protein 38.9 0.004
ath:AT3G62950 glutaredoxin family protein 37.0 0.014
ath:AT2G47870 glutaredoxin family protein 37.0 0.015
ath:AT2G30540 glutaredoxin family protein 36.6 0.018
pfa:PF07_0036 Cg6 protein 36.6 0.019
bbo:BBOV_II007430 18.m06616; thioredoxin-like protein 2 36.6
bbo:BBOV_II007360 18.m06610; Thioredoxin-like protein 2 36.6
tpv:TP01_0735 hypothetical protein 36.6 0.021
ath:AT3G15660 GRX4; GRX4 (GLUTAREDOXIN 4); metal ion binding 36.2 0.023
> tgo:TGME49_079400 glutaredoxin, putative ; K03676 glutaredoxin
3
Length=112
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 61/106 (57%), Positives = 77/106 (72%), Gaps = 2/106 (1%)
Query 18 MAPTSASE--VPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPF 75
MA T ASE V +W D L+ HK++VFSKS CPYC++AI A S+ DMHVE+IE +P+
Sbjct 1 MAETLASEAHVAEWADRLIKQHKVVVFSKSNCPYCRKAIEAFQSVKAKDMHVEEIEGSPY 60
Query 76 CDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
DAIQDY+K +TGARSVPRVFI G+F GG +DT+ GTL + L
Sbjct 61 MDAIQDYMKQQTGARSVPRVFIGGQFLGGAEDTIRAKADGTLVEKL 106
> tpv:TP01_0320 glutaredoxin-like protein; K03676 glutaredoxin
3
Length=151
Score = 127 bits (320), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 60/109 (55%), Positives = 76/109 (69%), Gaps = 1/109 (0%)
Query 8 SHFRTFSAFTMAPTSASEVPK-WVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMH 66
S F + +FT+ A + PK WVD LV HK++VFSKSYCPYC RA AL LN+ D+H
Sbjct 32 SDFLSKYSFTLPFKMAEKTPKDWVDSLVKKHKVVVFSKSYCPYCTRAKDALKKLNLHDLH 91
Query 67 VEQIENNPFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSG 115
VE++++NP D +QDYL TGARSVPRVF+NG+F+G TV V SG
Sbjct 92 VEELDSNPNMDQVQDYLNQLTGARSVPRVFVNGRFYGDSTKTVSDVESG 140
> cel:Y34D9A.6 glrx-10; GLutaRedoXin family member (glrx-10);
K03676 glutaredoxin 3
Length=105
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 55/95 (57%), Positives = 68/95 (71%), Gaps = 2/95 (2%)
Query 29 WVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIE--NNPFCDAIQDYLKTK 86
+VD L+ K++VFSKSYCPYC +A +AL S+NV ++ IE C+ IQDYL +
Sbjct 5 FVDGLLQSSKVVVFSKSYCPYCHKARAALESVNVKPDALQWIEIDERKDCNEIQDYLGSL 64
Query 87 TGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
TGARSVPRVFINGKFFGGGDDT G ++G L LL
Sbjct 65 TGARSVPRVFINGKFFGGGDDTAAGAKNGKLAALL 99
> pfa:PFC0271c GRX1, GRX-1; glutaredoxin 1; K03676 glutaredoxin
3
Length=111
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 55/108 (50%), Positives = 66/108 (61%), Gaps = 6/108 (5%)
Query 18 MAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNV-SDMHVEQIENNPFC 76
MA TS + V KWV+ +++ + I VF+K+ CPYC +AIS L N+ S MHVE IE NP
Sbjct 1 MAGTSEA-VKKWVNKIIEENIIAVFAKTECPYCIKAISILKGYNLNSHMHVENIEKNPDM 59
Query 77 DAIQDYLKTKTGARSVPRVFINGKFFGGGDDTV----EGVRSGTLQKL 120
IQ YLK TG SVPR+FIN GG DD V EG LQKL
Sbjct 60 ANIQAYLKELTGKSSVPRIFINKDVVGGCDDLVKENDEGKLKERLQKL 107
> bbo:BBOV_IV004320 23.m06161; glutaredoxin-like protein; K03676
glutaredoxin 3
Length=129
Score = 90.9 bits (224), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 65/99 (65%), Gaps = 0/99 (0%)
Query 23 ASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDY 82
S++ WV++ ++ K++VFSK+ CPYC +A L+S+ +D+ + Q+++NP I +Y
Sbjct 25 GSDISHWVNESINKSKVVVFSKTTCPYCIKANGILNSVAPNDLTIIQLDDNPDRAEIMEY 84
Query 83 LKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+ TGA +VPRVFI GKFFG TV SG L+K+L
Sbjct 85 FRETTGAATVPRVFIGGKFFGDCSKTVAANESGELKKVL 123
> tgo:TGME49_027150 glutaredoxin, putative
Length=332
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 41/93 (44%), Positives = 58/93 (62%), Gaps = 0/93 (0%)
Query 29 WVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTG 88
W+ + + HK++VF+ SYCPYC A+ L + V D+ I+ + IQD L+ TG
Sbjct 234 WIKEKIAKHKVVVFAMSYCPYCDTALEILRNAGVKDLGDVMIDRMDYTPQIQDILEEMTG 293
Query 89 ARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
AR+VPRVFI+G FFGG D E SG L+++L
Sbjct 294 ARTVPRVFIDGIFFGGCSDLEEAEASGKLKEIL 326
> ath:AT5G20500 glutaredoxin, putative; K03676 glutaredoxin 3
Length=135
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 48/108 (44%), Positives = 62/108 (57%), Gaps = 3/108 (2%)
Query 16 FTMAPTSASEVPK--WVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENN 73
F +SA+ P+ +V + HKI++FSKSYCPYC++A S L+ VE E
Sbjct 19 FISMVSSAASSPEADFVKKTISSHKIVIFSKSYCPYCKKAKSVFRELDQVPYVVELDERE 78
Query 74 PFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+IQ L G R+VP+VFINGK GG DDTV+ SG L KLL
Sbjct 79 DGW-SIQTALGEIVGRRTVPQVFINGKHLGGSDDTVDAYESGELAKLL 125
> ath:AT1G77370 glutaredoxin, putative
Length=130
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 44/112 (39%), Positives = 61/112 (54%), Gaps = 1/112 (0%)
Query 10 FRTFSAFTMAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQ 69
F + + +A+ V +V + + +KI++FSKSYCPYC R+ S L VE
Sbjct 17 FVVLCDLSNSAGAANSVSAFVQNAILSNKIVIFSKSYCPYCLRSKRIFSQLKEEPFVVE- 75
Query 70 IENNPFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
++ D IQ L G R+VP+VF+NGK GG DD + SG LQKLL
Sbjct 76 LDQREDGDQIQYELLEFVGRRTVPQVFVNGKHIGGSDDLGAALESGQLQKLL 127
> cpv:cgd2_2540 glutaredoxin related protein ; K03676 glutaredoxin
3
Length=108
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 59/97 (60%), Gaps = 1/97 (1%)
Query 21 TSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQ 80
T+ + + V+ + I V SKSYCPYC +AI++L S S + V QI+ IQ
Sbjct 6 TAMNSIKLLVESFISSGDICVISKSYCPYCIKAINSLKSAGYSPL-VMQIDGRVDTKEIQ 64
Query 81 DYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTL 117
DY + TG+ +VPRVF+ G+F GG DDT++ + G+L
Sbjct 65 DYCRELTGSGTVPRVFVKGRFIGGCDDTLKLLEDGSL 101
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/104 (40%), Positives = 60/104 (57%), Gaps = 1/104 (0%)
Query 18 MAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCD 77
M PT + V +L+D +++MVFSKS+CPYC R SSL +E E + D
Sbjct 1 MPPTGRDLLQARVKELIDSNRVMVFSKSFCPYCDRVKDLFSSLGAEYHSLELDECDDGSD 60
Query 78 AIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
IQ+ L+ TG ++VP VF+N GG D T++ + G+L KLL
Sbjct 61 -IQEALQELTGQKTVPNVFVNKTHVGGCDKTLQAHKDGSLAKLL 103
> cel:F10D7.3 hypothetical protein
Length=146
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 62/94 (65%), Gaps = 1/94 (1%)
Query 28 KWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKT 87
K V+D++ HK+MV+SK+YCP+ +R + L++ + DM + +++ + + +Q+ LK +
Sbjct 36 KIVNDVM-THKVMVYSKTYCPWSKRLKAILANYEIDDMKIVELDRSNQTEEMQEILKKYS 94
Query 88 GARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
G +VP++FI+GKF GG D+T G L+ LL
Sbjct 95 GRTTVPQLFISGKFVGGHDETKAIEEKGELRPLL 128
> dre:436677 glrx2, zgc:92698; glutaredoxin 2; K03676 glutaredoxin
3
Length=134
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/94 (39%), Positives = 57/94 (60%), Gaps = 1/94 (1%)
Query 28 KWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKT 87
++V D+V + +++FSK+ CPYC+ A + + + VE E+N +Q+ L T
Sbjct 18 QFVQDIVSSNCVVIFSKTTCPYCKMAKGVFNEIGATYKVVELDEHNDG-RRLQETLAELT 76
Query 88 GARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
GAR+VPRVFING+ GGG DT + + G L L+
Sbjct 77 GARTVPRVFINGQCIGGGSDTKQLHQQGKLLPLI 110
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 56/94 (59%), Gaps = 5/94 (5%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNV--SDMHVEQIENNPFCDAIQDYLKTKT 87
+ +L+D ++VFSKS+CP+C + LNV + + ++ +E+ QD L T
Sbjct 17 IKELIDSSAVVVFSKSFCPFCVKVKDLFKELNVKYNTIELDLMEDG---TNYQDLLHEMT 73
Query 88 GARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
G ++VP VFIN K GG D+T++ + G LQKLL
Sbjct 74 GQKTVPNVFINKKHIGGCDNTMKAHKDGVLQKLL 107
> hsa:2745 GLRX, GRX, GRX1, MGC117407; glutaredoxin (thioltransferase)
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=106
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 51/92 (55%), Gaps = 2/92 (2%)
Query 28 KWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVE--QIENNPFCDAIQDYLKT 85
++V+ + K++VF K CPYC+RA LS L + +E I + IQDYL+
Sbjct 4 EFVNCKIQPGKVVVFIKPTCPYCRRAQEILSQLPIKQGLLEFVDITATNHTNEIQDYLQQ 63
Query 86 KTGARSVPRVFINGKFFGGGDDTVEGVRSGTL 117
TGAR+VPRVFI GG D V +SG L
Sbjct 64 LTGARTVPRVFIGKDCIGGCSDLVSLQQSGEL 95
> mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase
3 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=652
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 58/92 (63%), Gaps = 5/92 (5%)
Query 32 DLVDGHKIMVFSKSYCPYCQRAISALSSLNV--SDMHVEQIENNPFCDAIQDYLKTKTGA 89
DL++G+++M+FSKSYCP+ R SSL V + + ++Q+++ ++Q+ L +
Sbjct 70 DLIEGNRVMIFSKSYCPHSTRVKELFSSLGVVYNILELDQVDDGA---SVQEVLTEISNQ 126
Query 90 RSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
++VP +F+N GG D T + ++G LQKLL
Sbjct 127 KTVPNIFVNKVHVGGCDRTFQAHQNGLLQKLL 158
> sce:YDR513W GRX2, TTR1; Cytoplasmic glutaredoxin, thioltransferase,
glutathione-dependent disulfide oxidoreductase involved
in maintaining redox state of target proteins, also exhibits
glutathione peroxidase activity, expression induced in
response to stress
Length=143
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/102 (35%), Positives = 56/102 (54%), Gaps = 3/102 (2%)
Query 23 ASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISAL-SSLNV--SDMHVEQIENNPFCDAI 79
+ E V DL+ ++ V +K+YCPYC+ +S L LNV S V +++ I
Sbjct 37 SQETVAHVKDLIGQKEVFVAAKTYCPYCKATLSTLFQELNVPKSKALVLELDEMSNGSEI 96
Query 80 QDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
QD L+ +G ++VP V+INGK GG D ++G L ++L
Sbjct 97 QDALEEISGQKTVPNVYINGKHIGGNSDLETLKKNGKLAEIL 138
> dre:449769 glrx, MGC113837, grx, wu:fc38f02, zgc:103707; glutaredoxin
(thioltransferase) (EC:1.8.4.1); K03676 glutaredoxin
3
Length=105
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 55/99 (55%), Gaps = 4/99 (4%)
Query 26 VPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCD--AIQDYL 83
+ ++V + K +VF K C YC A LS H E I+ + D +IQDYL
Sbjct 1 MAEFVKAQIKDGKGVVFCKPTCSYCILAKDVLSKYKFKAGHFELIDISARADMGSIQDYL 60
Query 84 KTKTGARSVPRVFINGKFFGGGDDTVEGV-RSGTLQKLL 121
+ TGAR+VPRVFI GGG D VEG+ RSG L+ +L
Sbjct 61 QQITGARTVPRVFIGEDCVGGGSD-VEGLDRSGKLEGML 98
> mmu:93692 Glrx, C86710, D13Wsu156e, Glrx1, Grx1, TTase; glutaredoxin
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=107
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 52/94 (55%), Gaps = 6/94 (6%)
Query 28 KWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIE----NNPFCDAIQDYL 83
++V+ + K++VF K CPYC++ LS L +E ++ NN AIQDYL
Sbjct 4 EFVNCKIQSGKVVVFIKPTCPYCRKTQEILSQLPFKQGLLEFVDITATNNT--SAIQDYL 61
Query 84 KTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTL 117
+ TGAR+VPRVFI GG D + ++G L
Sbjct 62 QQLTGARTVPRVFIGKDCIGGCSDLISMQQTGEL 95
> mmu:69367 Glrx2, 1700010P22Rik, AI645710, Grx2; glutaredoxin
2 (thioltransferase); K03676 glutaredoxin 3
Length=156
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGA 89
+ + + + +++FSK+ C YC A +NV+ VE ++ + + QD L TG
Sbjct 53 IQETISNNCVVIFSKTSCSYCSMAKKIFHDMNVNYKAVE-LDMLEYGNQFQDALHKMTGE 111
Query 90 RSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
R+VPR+F+NG+F GG DT + G L L+
Sbjct 112 RTVPRIFVNGRFIGGAADTHRLHKEGKLLPLV 143
> hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3
(EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=607
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 58/100 (58%), Gaps = 5/100 (5%)
Query 24 SEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNV--SDMHVEQIENNPFCDAIQD 81
E+ + + L++ ++++FSKSYCP+ R SSL V + + ++Q+++ +Q+
Sbjct 53 EELRRHLVGLIERSRVVIFSKSYCPHSTRVKELFSSLGVECNVLELDQVDDGA---RVQE 109
Query 82 YLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
L T ++VP +F+N GG D T + +SG LQKLL
Sbjct 110 VLSEITNQKTVPNIFVNKVHVGGCDQTFQAYQSGLLQKLL 149
> sce:YCL035C GRX1; Grx1p; K03676 glutaredoxin 3
Length=110
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 49/88 (55%), Gaps = 3/88 (3%)
Query 23 ASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISAL-SSLNV--SDMHVEQIENNPFCDAI 79
+ E K V DL+ ++I V SK+YCPYC A++ L L V S + V Q+ + I
Sbjct 3 SQETIKHVKDLIAENEIFVASKTYCPYCHAALNTLFEKLKVPRSKVLVLQLNDMKEGADI 62
Query 80 QDYLKTKTGARSVPRVFINGKFFGGGDD 107
Q L G R+VP ++INGK GG DD
Sbjct 63 QAALYEINGQRTVPNIYINGKHIGGNDD 90
> ath:AT5G63030 glutaredoxin, putative
Length=125
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 52/109 (47%), Gaps = 1/109 (0%)
Query 13 FSAFTMAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIEN 72
FS M+ V ++V + ++VFSK+YC YCQR L+ L + V +++
Sbjct 5 FSGNRMSKEEMEVVVNKAKEIVSAYPVVVFSKTYCGYCQRVKQLLTQLGAT-FKVLELDE 63
Query 73 NPFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
IQ L TG +VP VFI G GG D +E + G L LL
Sbjct 64 MSDGGEIQSALSEWTGQTTVPNVFIKGNHIGGCDRVMETNKQGKLVPLL 112
> xla:399044 glrx, MGC68461, grx, grx1; glutaredoxin (thioltransferase)
(EC:1.8.4.1); K03676 glutaredoxin 3
Length=107
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 29 WVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNP--FCDAIQDYLKTK 86
+V V K+ +F KS CP+C RA L+ + H+E I+ + F ++Q Y
Sbjct 5 FVQSKVKPSKVTMFEKSSCPFCVRAKGILTKYKFKEGHLEIIDISKLDFMSSLQQYFMQT 64
Query 87 TGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
TG +VPR++I K GG D V SG L+K L
Sbjct 65 TGESTVPRIYIGEKCIGGCSDLVPLENSGELEKAL 99
> hsa:51022 GLRX2, GRX2, bA101E13.1; glutaredoxin 2; K03676 glutaredoxin
3
Length=165
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/92 (32%), Positives = 50/92 (54%), Gaps = 1/92 (1%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGA 89
+ + + + +++FSK+ C YC A +NV + V +++ + + QD L TG
Sbjct 61 IQETISDNCVVIFSKTSCSYCTMAKKLFHDMNV-NYKVVELDLLEYGNQFQDALYKMTGE 119
Query 90 RSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
R+VPR+F+NG F GG DT + G L L+
Sbjct 120 RTVPRIFVNGTFIGGATDTHRLHKEGKLLPLV 151
> ath:AT4G28730 glutaredoxin family protein
Length=174
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 51/99 (51%), Gaps = 0/99 (0%)
Query 23 ASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDY 82
S + + + V + ++++SK++C YC + L V + VE + P +Q
Sbjct 66 GSRMEESIRKTVTENTVVIYSKTWCSYCTEVKTLFKRLGVQPLVVELDQLGPQGPQLQKV 125
Query 83 LKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
L+ TG +VP VF+ GK GG DTV+ R G L+ +L
Sbjct 126 LERLTGQHTVPNVFVCGKHIGGCTDTVKLNRKGDLELML 164
> ath:AT2G20270 glutaredoxin family protein
Length=179
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 48/92 (52%), Gaps = 0/92 (0%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGA 89
V V + ++V+SK++C Y + S SL V + VE + +Q+ L+ TG
Sbjct 78 VKTTVAENPVVVYSKTWCSYSSQVKSLFKSLQVEPLVVELDQLGSEGSQLQNVLEKITGQ 137
Query 90 RSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+VP VFI GK GG DT++ G L+ +L
Sbjct 138 YTVPNVFIGGKHIGGCSDTLQLHNKGELEAIL 169
> tgo:TGME49_027100 glutaredoxin, putative
Length=112
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query 33 LVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSV 92
++ HK++VF+K + P + L +N D+H+ + + C + LK KTG R
Sbjct 18 VIKSHKVVVFTKEHDPLSFEVVETLKQINPKDLHIVNL-DKCGCTQAEGLLKEKTGDRPG 76
Query 93 PRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
PRVFI+GKF DD ++ L+++L
Sbjct 77 PRVFIDGKFV---DDLETSLKDMDLREVL 102
> ath:AT5G40370 glutaredoxin, putative; K03676 glutaredoxin 3
Length=136
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 38/81 (46%), Gaps = 1/81 (1%)
Query 41 VFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFINGK 100
+ SK+YCPYC R L L VE ++ IQ L TG R+VP VFI G
Sbjct 42 ICSKTYCPYCVRVKELLQQLGAKFKAVE-LDTESDGSQIQSGLAEWTGQRTVPNVFIGGN 100
Query 101 FFGGGDDTVEGVRSGTLQKLL 121
GG D T + G L LL
Sbjct 101 HIGGCDATSNLHKDGKLVPLL 121
> ath:AT3G54900 CXIP1; CXIP1 (CAX INTERACTING PROTEIN 1); antiporter/
glutathione disulfide oxidoreductase
Length=173
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 51/95 (53%), Gaps = 9/95 (9%)
Query 30 VDDLVDGHKIMVFSKS-----YCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLK 84
++ LV+ K+++F K C + + L +LNV V +EN + ++ LK
Sbjct 75 LEKLVNSEKVVLFMKGTRDFPMCGFSNTVVQILKNLNVPFEDVNILEN----EMLRQGLK 130
Query 85 TKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQK 119
+ + P+++I G+FFGG D T+E ++G LQ+
Sbjct 131 EYSNWPTFPQLYIGGEFFGGCDITLEAFKTGELQE 165
> eco:b3610 grxC, ECK3600, JW3585, yibM; glutaredoxin 3; K03676
glutaredoxin 3
Length=83
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 45/83 (54%), Gaps = 4/83 (4%)
Query 39 IMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFIN 98
+ +++K CPYC RA + LSS VS + I+ N A ++ + ++G +VP++FI+
Sbjct 4 VEIYTKETCPYCHRAKALLSSKGVSFQELP-IDGNA---AKREEMIKRSGRTTVPQIFID 59
Query 99 GKFFGGGDDTVEGVRSGTLQKLL 121
+ GG DD G L LL
Sbjct 60 AQHIGGCDDLYALDARGGLDPLL 82
> hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=649
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 51/110 (46%), Gaps = 7/110 (6%)
Query 14 SAFTMAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSD--MHVEQIE 71
+ FT T+ S + +DGH +++FS+S C C SL V + ++Q E
Sbjct 45 AGFTSTATADSRA--LLQAYIDGHSVVIFSRSTCTRCTEVKKLFKSLCVPYFVLELDQTE 102
Query 72 NNPFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+ A++ L +P VF+ + GG T++ + G LQKLL
Sbjct 103 DG---RALEGTLSELAAETDLPVVFVKQRKIGGHGPTLKAYQEGRLQKLL 149
> ath:AT3G62960 glutaredoxin family protein
Length=102
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Query 39 IMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFIN 98
+++F+KS C C L V + +I+N+P C I+ L A +VP VF++
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALVRLGCANAVPAVFVS 71
Query 99 GKFFGGGDDTVEGVRSGTLQKLL 121
GK G +D + SG+L L+
Sbjct 72 GKLVGSTNDVMSLHLSGSLVPLI 94
> pfa:PFC0205c PfGLP-1; 1-cys-glutaredoxin-like protein-1; K07390
monothiol glutaredoxin
Length=171
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 53/98 (54%), Gaps = 10/98 (10%)
Query 30 VDDLVDGHKIMVF-----SKSYCPYCQRAISALSSLNVSD-MHVEQIENNPFCDAIQDYL 83
+ +L++ KI++F K C + ++ L+S+NV D ++++ ++NN +AI+ Y
Sbjct 77 IKELLEQEKIVLFMKGTPEKPLCGFSANVVNILNSMNVKDYVYIDVMKNNNLREAIKIY- 135
Query 84 KTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+ +P +++N F GG D + G L+K++
Sbjct 136 ---SNWPYIPNLYVNNNFIGGYDIISDLYNRGELEKII 170
> sce:YLR364W GRX8; Glutaredoxin that employs a dithiol mechanism
of catalysis; monomeric; activity is low and null mutation
does not affect sensitivity to oxidative stress; GFP-fusion
protein localizes to the cytoplasm; expression strongly induced
by arsenic
Length=109
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 45/98 (45%), Gaps = 11/98 (11%)
Query 31 DDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDM-------HVEQIENNPFCDAIQDYL 83
++++ H S S+CP C A S + LNV D + + E + A Q +
Sbjct 9 EEMIKSHPYFQLSASWCPDCVYANSIWNKLNVQDKVFVFDIGSLPRNEQEKWRIAFQKVV 68
Query 84 KTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
G+R++P + +NGKF+G GTL++ L
Sbjct 69 ----GSRNLPTIVVNGKFWGTESQLHRFEAKGTLEEEL 102
> cel:C07G1.8 glrx-22; GLutaRedoXin family member (glrx-22)
Length=131
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 48/97 (49%), Gaps = 9/97 (9%)
Query 14 SAFTMAPTSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENN 73
SA T + SE K + + V H +++++K C YC + A + L +H + N
Sbjct 4 SASTPPKPTLSEHSKKIVEEVKEHAVVLYTKDGCGYC---VKAKNELYEDGIHYTEKNLN 60
Query 74 PFCDAI---QDYLK---TKTGARSVPRVFINGKFFGG 104
I Q+Y++ T R+VP++FI GKF GG
Sbjct 61 TVSKVIPNPQEYIQGLMDLTRQRTVPQIFICGKFVGG 97
> dre:569013 MGC152951; zgc:152951
Length=372
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 19/91 (20%)
Query 38 KIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDA-------IQDYLKTKTGAR 90
++ V+S CP+C RA + L +L V P CD I+ +K TG
Sbjct 11 RVTVYSVPGCPHCTRAKTTLGALGV-----------PVCDVDVNKHREIRALVKELTGHS 59
Query 91 SVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
SVP++F N + G +D ++ + L+ LL
Sbjct 60 SVPQIFFNNLYVGNNED-LQNLDPKRLEHLL 89
> sce:YDL010W GRX6; Cis-golgi localized monothiol glutaredoxin
that binds an iron-sulfur cluster; more similar in activity
to dithiol than other monothiol glutaredoxins; involved in
the oxidative stress response; functional overlap with GRX7
Length=231
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 47/89 (52%), Gaps = 2/89 (2%)
Query 21 TSASEVPKWVDDLVDGHKIMVFSKSYCPYCQRAISALSSLN--VSDMHVEQIENNPFCDA 78
T A V K ++D I++FSKS C Y + L + + + ++ +++ + +
Sbjct 110 TKAFNVQKEYSLILDLSPIIIFSKSTCSYSKGMKELLENEYQFIPNYYIIELDKHGHGEE 169
Query 79 IQDYLKTKTGARSVPRVFINGKFFGGGDD 107
+Q+Y+K TG +VP + +NG GG ++
Sbjct 170 LQEYIKLVTGRGTVPNLLVNGVSRGGNEE 198
> ath:AT2G47880 glutaredoxin family protein
Length=102
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 41/83 (49%), Gaps = 1/83 (1%)
Query 39 IMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFIN 98
+++F+KS C C L V + +I+N+P C I+ L + +VP VF+
Sbjct 13 VVIFTKSSCCLCYAVQILFRDLRVQPT-IHEIDNDPDCREIEKALLRLGCSTAVPAVFVG 71
Query 99 GKFFGGGDDTVEGVRSGTLQKLL 121
GK G ++ + SG+L L+
Sbjct 72 GKLVGSTNEVMSLHLSGSLVPLI 94
> sce:YBR014C GRX7; Cis-golgi localized monothiol glutaredoxin;
more similar in activity to dithiol than other monothiol glutaredoxins;
involved in the oxidative stress response; does
not bind metal ions; functional overlap with GRX6
Length=203
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 40/74 (54%), Gaps = 2/74 (2%)
Query 39 IMVFSKSYCPYCQRAISALSSLNV--SDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVF 96
++VFSK+ CPY ++ + L++ +V +++ + +QD ++ TG R+VP V
Sbjct 100 MIVFSKTGCPYSKKLKALLTNSYTFSPSYYVVELDRHEHTKELQDQIEKVTGRRTVPNVI 159
Query 97 INGKFFGGGDDTVE 110
I G GG + E
Sbjct 160 IGGTSRGGYTEIAE 173
> tpv:TP02_0196 hypothetical protein
Length=177
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 55/108 (50%), Gaps = 9/108 (8%)
Query 15 AFTMAPTSASEVPKWVDDLVDGHKIMVFSK--SYCPYCQRAISALSSLNVSDMHVEQIEN 72
AF T+ ++ + L+ H +++F K P+C+ + + ++ LN S + E
Sbjct 68 AFKTRETTLDKIKR----LLREHPVLLFMKGDKAEPFCRFSRAVVNMLNSSGVRFEGY-- 121
Query 73 NPFCDA-IQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQK 119
N F D +++ LKT + + P++++NG GG D E + TL+K
Sbjct 122 NIFDDPNLREELKTYSNWPTYPQLYVNGNLIGGHDIIKELYETNTLRK 169
> ath:AT2G38270 CXIP2; CXIP2 (CAX-INTERACTING PROTEIN 2); electron
carrier/ protein disulfide oxidoreductase
Length=293
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 44/97 (45%), Gaps = 6/97 (6%)
Query 30 VDDLVDGHKIMVF-----SKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLK 84
+D LV K++ F S C + QR + L S V D + ++ + +++ LK
Sbjct 197 IDRLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGV-DYETVDVLDDEYNHGLRETLK 255
Query 85 TKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+ + P++F+ G+ GG D +G L +L
Sbjct 256 NYSNWPTFPQIFVKGELVGGCDILTSMYENGELANIL 292
> ath:AT4G04950 thioredoxin family protein
Length=488
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 11/98 (11%)
Query 30 VDDLVDGHKIMVFSKSYC--PYCQRAISALSSLNVSDMHVEQIENNPFC----DAIQDYL 83
++ LV+ +M+F K P C S V ++ E+IE F D ++ L
Sbjct 287 LEGLVNSKPVMLFMKGRPEEPKC-----GFSGKVVEILNQEKIEFGSFDILLDDEVRQGL 341
Query 84 KTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
K + S P++++ G+ GG D +E +SG L+K+L
Sbjct 342 KVYSNWSSYPQLYVKGELMGGSDIVLEMQKSGELKKVL 379
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 22/103 (21%), Positives = 49/103 (47%), Gaps = 9/103 (8%)
Query 21 TSASEVPKWVDDLVDGHKIMVFSKSY-----CPYCQRAISALSSLNVSDMHVEQIENNPF 75
++A + ++ L + H +M+F K C + ++ + L +NV + + +N
Sbjct 148 STADALKSRLEKLTNSHPVMLFMKGIPEEPRCGFSRKVVDILKEVNVDFGSFDILSDN-- 205
Query 76 CDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQ 118
+++ LK + + P+++ NG+ GG D + SG L+
Sbjct 206 --EVREGLKKFSNWPTFPQLYCNGELLGGADIAIAMHESGELK 246
> ath:AT3G62950 glutaredoxin family protein
Length=103
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 21/93 (22%), Positives = 44/93 (47%), Gaps = 2/93 (2%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGA 89
+ DL ++F+KS C C + L S + +++ +P ++ L+ +
Sbjct 4 IRDLSSKKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPEGREMERALRALGSS 62
Query 90 R-SVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+VP VF+ G++ G D + G+L+++L
Sbjct 63 NPAVPAVFVGGRYIGSAKDIISFHVDGSLKQML 95
> ath:AT2G47870 glutaredoxin family protein
Length=103
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 44/93 (47%), Gaps = 2/93 (2%)
Query 30 VDDLVDGHKIMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGA 89
V DL ++F+KS C C + L S + +++ +P ++ L G+
Sbjct 4 VRDLASEKAAVIFTKSSCCMCHSIKTLFYELGASPA-IHELDKDPQGPDMERALFRVFGS 62
Query 90 R-SVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+VP VF+ G++ G D + G+L+++L
Sbjct 63 NPAVPAVFVGGRYVGSAKDVISFHVDGSLKQML 95
> ath:AT2G30540 glutaredoxin family protein
Length=102
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Query 39 IMVFSKSYCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFIN 98
+++FSKS C L V V +I+ +P C I+ L + VP +F+
Sbjct 13 VVIFSKSSCCMSYAVQVLFQDLGVHPT-VHEIDKDPECREIEKALMRLGCSTPVPAIFVG 71
Query 99 GKFFGGGDDTVEGVRSGTLQKLL 121
GK G ++ + SG+L L+
Sbjct 72 GKLIGSTNEVMSLHLSGSLVPLV 94
> pfa:PF07_0036 Cg6 protein
Length=272
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Query 46 YCPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTGARSVPRVFINGKFFGGG 105
YC Y ++AI L V +H I +N +++ LK + + P++++N KF GG
Sbjct 200 YCKYSKQAIHILKLNKVKQIHTVNILDNQ---ELRNALKIYSNWPTFPQLYVNQKFIGGI 256
Query 106 DDTVE 110
D E
Sbjct 257 DKLQE 261
> bbo:BBOV_II007430 18.m06616; thioredoxin-like protein 2
Length=202
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 46/89 (51%), Gaps = 5/89 (5%)
Query 33 LVDGHKIMVFSKSYC--PYCQRAISALSSLNVSDMHVEQIENNPFCD-AIQDYLKTKTGA 89
L+ H+I++F K P+C+ + + ++ LN S VE N F D +++ LK +
Sbjct 114 LISTHRILLFMKGEKSDPFCRYSKAVVNMLNESG--VEYDTYNIFEDPELREELKVYSNW 171
Query 90 RSVPRVFINGKFFGGGDDTVEGVRSGTLQ 118
+ P++++NG GG D E +L+
Sbjct 172 PTYPQLYVNGSLIGGHDIIKELYEQNSLR 200
> bbo:BBOV_II007360 18.m06610; Thioredoxin-like protein 2
Length=202
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 45/88 (51%), Gaps = 5/88 (5%)
Query 22 SASEVPKWVDDLVDGHKIMVFSKSYC--PYCQRAISALSSLNVSDMHVEQIENNPFCD-A 78
S + + L+ H+I++F K P+C+ + + ++ LN S VE N F D
Sbjct 103 SGESTQERIKRLISTHRILLFMKGEKSDPFCRYSKAVVNMLNESG--VEYDTYNIFEDPE 160
Query 79 IQDYLKTKTGARSVPRVFINGKFFGGGD 106
+++ LK + + P++++NG GG D
Sbjct 161 LREELKVYSNWPTYPQLYVNGSLIGGHD 188
> tpv:TP01_0735 hypothetical protein
Length=193
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 19/93 (20%), Positives = 49/93 (52%), Gaps = 8/93 (8%)
Query 34 VDGHKIMVFSKSY-----CPYCQRAISALSSLNVSDMHVEQIENNPFCDAIQDYLKTKTG 88
++ +++++F K C + ++A+ L +L + + + + + +++ LK +
Sbjct 101 IETYEVVLFMKGTARKPACGFSKQALDILKALKIDTIRTVNVLED---EDVRNGLKAYSK 157
Query 89 ARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+P++++ GKF GG + + G+L+KLL
Sbjct 158 YPYIPQLYVRGKFVGGLEKITKMFSDGSLEKLL 190
> ath:AT3G15660 GRX4; GRX4 (GLUTAREDOXIN 4); metal ion binding
Length=169
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 25/109 (22%), Positives = 48/109 (44%), Gaps = 9/109 (8%)
Query 18 MAPTSASEVPKWVDDLVDGHKIMVFSKSY-----CPYCQRAISALSSLNVSDMHVEQIEN 72
+ P S + V++ V + +M++ K C + A+ L NV +E+
Sbjct 57 VPPDSTDSLKDIVENDVKDNPVMIYMKGVPESPQCGFSSLAVRVLQQYNVPISSRNILED 116
Query 73 NPFCDAIQDYLKTKTGARSVPRVFINGKFFGGGDDTVEGVRSGTLQKLL 121
+A++ + T P++FI G+F GG D + + G L++ L
Sbjct 117 QELKNAVKSFSHWPT----FPQIFIKGEFIGGSDIILNMHKEGELEQKL 161
Lambda K H
0.321 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40