bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0739_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
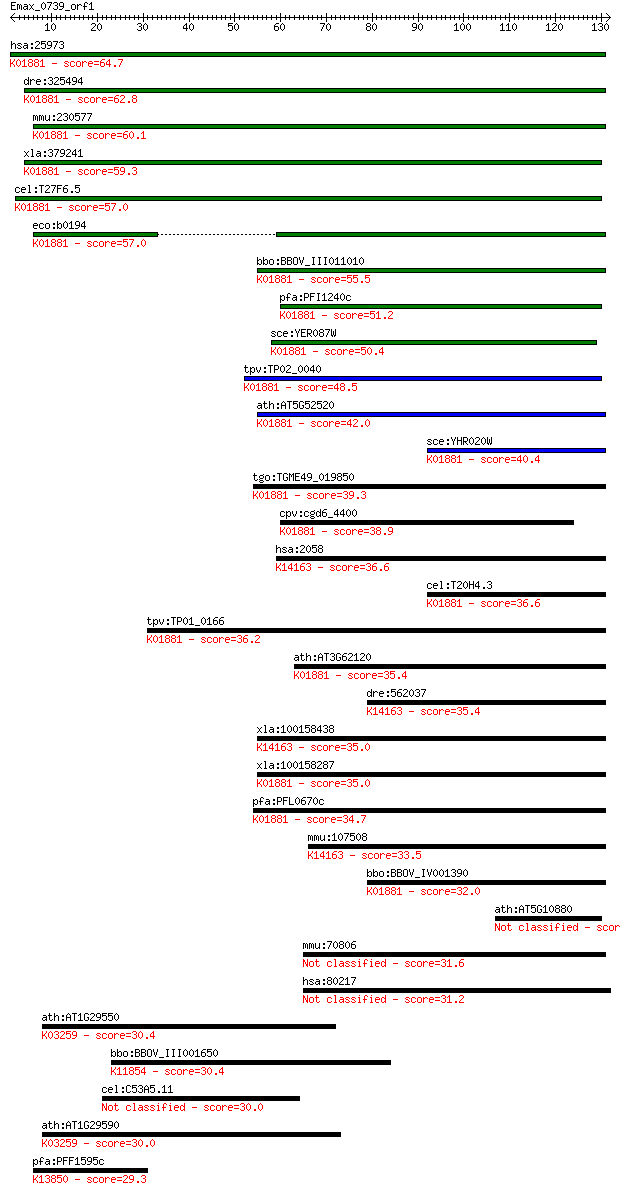
hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS; p... 64.7 8e-11
dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA ... 62.8 2e-10
mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase (... 60.1 2e-09
xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochon... 59.3 3e-09
cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2... 57.0 1e-08
eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase ... 57.0 1e-08
bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core do... 55.5 4e-08
pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15); K0... 51.2 8e-07
sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA sy... 50.4 1e-06
tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 48.5 6e-06
ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / ami... 42.0 5e-04
sce:YHR020W Protein of unknown function that may interact with... 40.4 0.002
tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.1... 39.3 0.003
cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA... 38.9 0.004
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 36.6 0.018
cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1... 36.6 0.019
tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synth... 36.2 0.030
ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family ... 35.4 0.041
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 35.4 0.048
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 35.0 0.059
xla:100158287 hypothetical protein LOC100158287; K01881 prolyl... 35.0 0.062
pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative ... 34.7 0.079
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 33.5 0.19
bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.... 32.0 0.55
ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related 31.6 0.61
mmu:70806 Wdr96, 4632415N18Rik, 4930428C11Rik, 4930463G05Rik, ... 31.6 0.67
hsa:80217 WDR96, C10orf79, FLJ22944, FLJ36006, bA373N18.2; WD ... 31.2 0.79
ath:AT1G29550 eukaryotic translation initiation factor 4E, put... 30.4 1.5
bbo:BBOV_III001650 17.m07166; ubiquitin carboxyl-terminal hydr... 30.4 1.6
cel:C53A5.11 hypothetical protein 30.0 2.0
ath:AT1G29590 eukaryotic translation initiation factor 4E, put... 30.0 2.0
pfa:PFF1595c VAR; erythrocyte membrane protein 1, PfEMP1; K138... 29.3 2.9
> hsa:25973 PARS2, DKFZp727A071, MGC14416, MGC19467, MT-PRORS;
prolyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=475
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 43/141 (30%), Positives = 66/141 (46%), Gaps = 15/141 (10%)
Query 1 PVYRFQADPGTMGGPCSHEYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQ------ 54
P + QAD GT+GG SHE+ + +G DR+ + P A+ E ++ Q
Sbjct 240 PFVKVQADVGTIGGTVSHEFQLPVDIGEDRL--AICP--RCSFSANMETLDLSQMNCPAC 295
Query 55 -----NSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVH 109
+ +EVGH F L Y +FT+ G + M YGLGV+R+LA V
Sbjct 296 QGPLTKTKGIEVGHTFYLGTKYSSIFNAQFTNVCGKPTLAEMGCYGLGVTRILAAAIEVL 355
Query 110 QDSKGLLFPPQVAPFCAAILP 130
+ +P +AP+ A ++P
Sbjct 356 STEDCVRWPSLLAPYQACLIP 376
> dre:325494 pars2, fc83e06, wu:fc83e06, zgc:77163; prolyl-tRNA
synthetase (mitochondrial) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=477
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 63/135 (46%), Gaps = 8/135 (5%)
Query 4 RFQADPGTMGGPCSHEYHVSSILGSDRIV--------SGVGPLNESKKDASAEDMEYHQN 55
+ QAD G +GG SHE+ + + +G D+++ + + + + S
Sbjct 244 QVQADTGNIGGTLSHEFQLPADIGEDKLLVCGNCGFSANIETMEPGQAKCSQCQTGKLTE 303
Query 56 SSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGL 115
S +EVGH F L Y +A + F ++ V M +GLGV+R+LA V +
Sbjct 304 SRGIEVGHTFYLGTKYSQAFKALFVNASNVPAVAEMGCFGLGVTRILAASIEVMSTEDAI 363
Query 116 LFPPQVAPFCAAILP 130
+P +AP+ +LP
Sbjct 364 HWPGLIAPYQVCVLP 378
> mmu:230577 Pars2, BC027073, MGC47008; prolyl-tRNA synthetase
(mitochondrial)(putative) (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=511
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 64/136 (47%), Gaps = 15/136 (11%)
Query 6 QADPGTMGGPCSHEYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQN---------- 55
+AD G++GG SHE+ + +G DR+V V P A+ E ++ Q
Sbjct 281 RADVGSIGGTMSHEFQLPVDIGEDRLV--VCP--SCHFSANTEILDLSQKICPDCQGPLT 336
Query 56 -SSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKG 114
+ +EVGH F L Y FT++ G + M YGLGV+R+LA V
Sbjct 337 ETKGIEVGHTFYLSTKYSSIFNALFTNAHGESLLAEMGCYGLGVTRILAAAIEVLSTEDC 396
Query 115 LLFPPQVAPFCAAILP 130
+ +P +AP+ ++P
Sbjct 397 IRWPRLLAPYQVCVIP 412
> xla:379241 pars2, MGC53152; prolyl-tRNA synthetase 2, mitochondrial
(putative); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=466
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 38/134 (28%), Positives = 65/134 (48%), Gaps = 9/134 (6%)
Query 4 RFQADPGTMGGPCSHEYHVSSILGSDRIV--------SGVGPLNESKKDASAEDMEYHQN 55
+ QAD G +GG SHE+H+ + +G DR++ + V L +K+ + +
Sbjct 234 KVQADTGNIGGKMSHEFHLPANIGEDRLLVCGSCDFSANVELLLHDEKNCPVCTGQL-KE 292
Query 56 SSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGL 115
+ +E+GH F L Y + DS+ + M YG+GVSRLLA V + +
Sbjct 293 TKGIEIGHTFYLGTKYSHVFKANCYDSQKTPFLAEMGCYGIGVSRLLAASLEVLSNEDDI 352
Query 116 LFPPQVAPFCAAIL 129
+P +AP+ ++
Sbjct 353 HWPGLIAPYQVCLI 366
> cel:T27F6.5 prs-2; Prolyl tRNA Synthetase family member (prs-2);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=454
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 47/142 (33%), Positives = 67/142 (47%), Gaps = 23/142 (16%)
Query 2 VYRFQADPGTMGGPCSHEYHVSSILGSDRI-----------VSGVGPLNESKKDASAED- 49
V + +AD G GG SHEYH+ S L D + G GP +K + AE+
Sbjct 196 VIKVEADSGVHGGHISHEYHLKSSLEEDFVNFCENCKNHNKFEGPGP---AKCEKCAENS 252
Query 50 MEYHQNSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPL-MNSYGLGVSRLL-AHLAL 107
+ Q ++E+ H F L Y +A +F PL M +G+GV+RLL A + L
Sbjct 253 SKTVQKIPSVEIAHTFHLGTKYSEALGAKFQGK------PLDMCCFGIGVTRLLPAAIDL 306
Query 108 VHQDSKGLLFPPQVAPFCAAIL 129
+ K L P +APF A I+
Sbjct 307 LSVSDKALRLPRAIAPFDAVII 328
> eco:b0194 proS, drpA, ECK0194, JW0190; prolyl-tRNA synthetase
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=572
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 59 LEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFP 118
+EVGH FQL Y +A + G ++ M YG+GV+R++A + D +G+++P
Sbjct 409 IEVGHIFQLGTKYSEALKASVQGEDGRNQILTMGCYGIGVTRVVAAAIEQNYDERGIVWP 468
Query 119 PQVAPFCAAILP 130
+APF AILP
Sbjct 469 DAIAPFQVAILP 480
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 6 QADPGTMGGPCSHEYHVSSILGSDRIV 32
QAD G++GG SHE+ V + G D +V
Sbjct 196 QADTGSIGGSASHEFQVLAQSGEDDVV 222
> bbo:BBOV_III011010 17.m07948; tRNA synthetase class II core
domain containing protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=439
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 30/78 (38%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Query 55 NSSTL-EVGHCFQLPDTYCKAARTRFTDSRGALRVPL-MNSYGLGVSRLLAHLALVHQDS 112
N TL E+ H FQL D A ++ + G ++ P+ +NSYG+G+ RLL A H DS
Sbjct 232 NKKTLMEIAHIFQLGDAITTEAGLKY-EGEGRMKHPVYLNSYGIGIHRLLQAAASQHADS 290
Query 113 KGLLFPPQVAPFCAAILP 130
+G+ P +AP+ A++P
Sbjct 291 EGIRLPQIIAPYDVAVIP 308
> pfa:PFI1240c prolyl-t-RNA synthase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=579
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 36/70 (51%), Gaps = 0/70 (0%)
Query 60 EVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPP 119
E H F+L D Y ++ D + + LM SYG+G+ RLL L D +G+ P
Sbjct 299 EAAHIFKLGDYYSNKLDIKYLDKKNEKKNILMGSYGIGIYRLLYFLIENFYDEEGIKLPQ 358
Query 120 QVAPFCAAIL 129
QVAPF ++
Sbjct 359 QVAPFSVYLI 368
> sce:YER087W AIM10; Aim10p (EC:6.1.1.15); K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=576
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 58 TLEVGHCFQLPDTYCKAARTRFTDSRGALRVPL-MNSYGLGVSRLLAHLALVHQDSKGLL 116
++EVGH F L + Y K +F D + M YG+GVSRL+ +A + +DS G
Sbjct 378 SIEVGHIFLLGNKYSKPLNVKFVDKENKNETFVHMGCYGIGVSRLVGAIAELGRDSNGFR 437
Query 117 FPPQVAPFCAAI 128
+P +AP+ +I
Sbjct 438 WPAIMAPYKVSI 449
> tpv:TP02_0040 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=461
Score = 48.5 bits (114), Expect = 6e-06, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query 52 YHQNSST-LEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQ 110
+ QNS+T LEVGH F+ Y +F S MNSYG+G++RLL L +
Sbjct 256 HDQNSNTELEVGHLFKFGTHYSDVYGLKFEISGRKREKLYMNSYGIGINRLLNVLINNYS 315
Query 111 DSKGLLFPPQVAPFCAAIL 129
D G+ P +AP+ I+
Sbjct 316 DEFGIKLPQLIAPYDVTII 334
> ath:AT5G52520 OVA6; OVA6 (OVULE ABORTION 6); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / proline-tRNA ligase;
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=543
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query 55 NSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKG 114
+ L+ G L + +A T+F D G + S+ + +R + + + H D G
Sbjct 279 DRKALQAGTSHNLGQNFSRAFGTQFADENGERQHVWQTSWAVS-TRFVGGIIMTHGDDTG 337
Query 115 LLFPPQVAPFCAAILP 130
L+ PP++AP I+P
Sbjct 338 LMLPPKIAPIQVVIVP 353
> sce:YHR020W Protein of unknown function that may interact with
ribosomes, based on co-purification experiments; has similarity
to proline-tRNA ligase; YHR020W is an essential gene
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=688
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 29/39 (74%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+GL +R++ + ++H D+KGL+ PP+V+ F + ++P
Sbjct 443 NSWGLS-TRVIGVMVMIHSDNKGLVIPPRVSQFQSVVIP 480
> tgo:TGME49_019850 prolyl-tRNA synthetase, putative (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=711
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 34/77 (44%), Gaps = 1/77 (1%)
Query 54 QNSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSK 113
+N ++ L + K F D G R+ S+G +R L + + H D K
Sbjct 430 ENGRGIQAATSHLLGTNFAKMFEIEFEDEEGHKRLVHQTSWGC-TTRSLGVMIMTHGDDK 488
Query 114 GLLFPPQVAPFCAAILP 130
GL+ PP+VA I+P
Sbjct 489 GLVIPPRVASVQVVIIP 505
> cpv:cgd6_4400 proline-tRNA synthetase; class II aaRS (ybak RNA
binding domain plus tRNA synthetase) ; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=719
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 33/70 (47%), Gaps = 7/70 (10%)
Query 60 EVGHCFQ------LPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSK 113
E+G Q L + K F D +G S+GL +R + + + H D+K
Sbjct 441 EIGRAVQAATSHLLGQNFSKMFGVEFEDEKGNKEYAHQTSWGL-TTRAIGVMIMTHGDNK 499
Query 114 GLLFPPQVAP 123
GL+ PP+VAP
Sbjct 500 GLVLPPKVAP 509
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 36/74 (48%), Gaps = 3/74 (4%)
Query 59 LEVGHCFQLPDTYCKAARTRFTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDSKGLL 116
++ G L + K F D + G + NS+GL +R + + +VH D+ GL+
Sbjct 1236 IQGGTSHHLGQNFSKMFEIVFEDPKIPGEKQFAYQNSWGL-TTRTIGVMTMVHGDNMGLV 1294
Query 117 FPPQVAPFCAAILP 130
PP+VA I+P
Sbjct 1295 LPPRVACVQVVIIP 1308
> cel:T20H4.3 prs-1; Prolyl tRNA Synthetase family member (prs-1);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=581
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 92 NSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
NS+GL +R + + ++H D KGL+ PP+VA ++P
Sbjct 346 NSWGLS-TRTIGAMVMIHGDDKGLVLPPRVAAVQVIVVP 383
> tpv:TP01_0166 prolyl-tRNA synthetase; K01881 prolyl-tRNA synthetase
[EC:6.1.1.15]
Length=683
Score = 36.2 bits (82), Expect = 0.030, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 44/104 (42%), Gaps = 6/104 (5%)
Query 31 IVSGVGPLNE----SKKDASAEDMEYHQNSSTLEVGHCFQLPDTYCKAARTRFTDSRGAL 86
++ GV NE SK S E N ++ L T+ ++ D G
Sbjct 390 VIKGVKSENEKFAGSKMTTSLEAF-VPANGRGVQAATSHLLGTTFSDMFDIKYEDEEGEK 448
Query 87 RVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
R S+G +R + + +VH D +GL+ PP+VA I+P
Sbjct 449 RKVHQTSWGF-TTRSIGIMIMVHGDDRGLVVPPRVAKTQVVIVP 491
> ath:AT3G62120 tRNA synthetase class II (G, H, P and S) family
protein; K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=530
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Query 63 HCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVA 122
HC L + K F + + + NS+ +R + + + H D KGL+ PP+VA
Sbjct 275 HC--LGQNFAKMFEINFENEKAETEMVWQNSWAYS-TRTIGVMIMTHGDDKGLVLPPKVA 331
Query 123 PFCAAILP 130
++P
Sbjct 332 SVQVVVIP 339
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 35.4 bits (80), Expect = 0.048, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 79 FTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
F D + G ++ NS+G+ +R + L +VH D+ GL+ PP+VA I+P
Sbjct 1255 FEDPKKPGEKQLAYQNSWGI-TTRTIGVLTMVHGDNMGLVLPPRVACLQVIIIP 1307
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 35.0 bits (79), Expect = 0.059, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 37/78 (47%), Gaps = 3/78 (3%)
Query 55 NSSTLEVGHCFQLPDTYCKAARTRFTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDS 112
+S ++ L + K F D + G + NS+GL +R + + +VH D+
Sbjct 1219 SSRAIQGATSHHLGQNFSKMFEIVFEDPKTPGLKQYAYQNSWGLS-TRTIGAMVMVHADN 1277
Query 113 KGLLFPPQVAPFCAAILP 130
G++ PP+VA I+P
Sbjct 1278 TGMVLPPRVACVQIIIIP 1295
> xla:100158287 hypothetical protein LOC100158287; K01881 prolyl-tRNA
synthetase [EC:6.1.1.15]
Length=655
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 37/78 (47%), Gaps = 3/78 (3%)
Query 55 NSSTLEVGHCFQLPDTYCKAARTRFTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDS 112
+S ++ L + K F D + G + NS+GL +R + + +VH D+
Sbjct 375 SSRAIQGATSHHLGQNFSKIFEIVFEDPKTPGLKQYAYQNSWGLS-TRTIGAMVMVHADN 433
Query 113 KGLLFPPQVAPFCAAILP 130
G++ PP+VA I+P
Sbjct 434 IGMVLPPRVACVQVVIIP 451
> pfa:PFL0670c bifunctional aminoacyl-tRNA synthetase, putative
(EC:6.1.1.15); K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=746
Score = 34.7 bits (78), Expect = 0.079, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 33/77 (42%), Gaps = 1/77 (1%)
Query 54 QNSSTLEVGHCFQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSK 113
+N ++ L + K + F D + S+G +R + + + H D K
Sbjct 469 ENGRAIQAATSHYLGTNFAKMFKIEFEDENEVKQYVHQTSWGC-TTRSIGIMIMTHGDDK 527
Query 114 GLLFPPQVAPFCAAILP 130
GL+ PP V+ + I+P
Sbjct 528 GLVLPPNVSKYKVVIVP 544
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Query 66 QLPDTYCKAARTRFTDSR--GALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAP 123
L + K F D + G + S+GL +R + + +VH D+ GL+ PP+VA
Sbjct 1243 HLGQNFSKMCEIVFEDPKTPGEKQFAYQCSWGL-TTRTIGVMVMVHGDNMGLVLPPRVAS 1301
Query 124 FCAAILP 130
++P
Sbjct 1302 VQVVVIP 1308
> bbo:BBOV_IV001390 21.m02889; prolyl-tRNA synthetase (EC:6.1.1.15);
K01881 prolyl-tRNA synthetase [EC:6.1.1.15]
Length=705
Score = 32.0 bits (71), Expect = 0.55, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 79 FTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPFCAAILP 130
F D G ++ S+G +R + ++H D +GL+ PP+V+ I+P
Sbjct 456 FEDENGVKQLAHQTSWGF-TTRSIGVSIMIHGDDQGLVIPPRVSKVQIVIVP 506
> ath:AT5G10880 tRNA synthetase-related / tRNA ligase-related
Length=309
Score = 31.6 bits (70), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 107 LVHQDSKGLLFPPQVAPFCAAIL 129
+ H D KGL+FPP+VAP ++
Sbjct 89 MTHGDDKGLVFPPKVAPVQVVVI 111
> mmu:70806 Wdr96, 4632415N18Rik, 4930428C11Rik, 4930463G05Rik,
AI429486, D19Ertd652e; WD repeat domain 96
Length=1682
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 28/66 (42%), Gaps = 0/66 (0%)
Query 65 FQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPF 124
F LP + F+D RG L+ L + Y V L+ L SK F QV
Sbjct 570 FTLPVMLPEVVPENFSDERGRLKDDLTHKYLYEVEHTLSSAVLGFTGSKIFGFCSQVPYI 629
Query 125 CAAILP 130
C+ ++P
Sbjct 630 CSYVMP 635
> hsa:80217 WDR96, C10orf79, FLJ22944, FLJ36006, bA373N18.2; WD
repeat domain 96
Length=1665
Score = 31.2 bits (69), Expect = 0.79, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 65 FQLPDTYCKAARTRFTDSRGALRVPLMNSYGLGVSRLLAHLALVHQDSKGLLFPPQVAPF 124
F LP T T F D RG L+ +++ Y + L+ L Q ++ F QV
Sbjct 554 FTLP-TLLPQVSTTFADERGRLKDEIIHKYLYELEHALSSAVLGFQSNQIYGFCSQVPYI 612
Query 125 CAAILPQ 131
C+ +LP+
Sbjct 613 CSYLLPE 619
> ath:AT1G29550 eukaryotic translation initiation factor 4E, putative
/ eIF-4E, putative / eIF4E, putative / mRNA cap-binding
protein, putative; K03259 translation initiation factor
4E
Length=240
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 12/66 (18%)
Query 8 DPGTMGGPCSHEYHVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSST--LEVGHCF 65
DP T P E HVS+I + +SG +K S E Y ST ++ HCF
Sbjct 18 DPNTTTSPSPKEKHVSAI----KAISG------DEKAPSKEKKNYASKKSTTVIQKSHCF 67
Query 66 QLPDTY 71
Q T+
Sbjct 68 QNSWTF 73
> bbo:BBOV_III001650 17.m07166; ubiquitin carboxyl-terminal hydrolase
family protein; K11854 ubiquitin carboxyl-terminal hydrolase
35/38 [EC:3.1.2.15]
Length=1073
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 3/62 (4%)
Query 23 SSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSSTL-EVGHCFQLPDTYCKAARTRFTD 81
SSI G+ RI+S V + +A +++ + Q + E+ H LPDTY + + D
Sbjct 85 SSIPGNSRIISTVDTTKRADDNAKSDEYRHFQEIPAVDEIVHL--LPDTYSEKLMIQLID 142
Query 82 SR 83
R
Sbjct 143 DR 144
> cel:C53A5.11 hypothetical protein
Length=430
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 11/43 (25%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 21 HVSSILGSDRIVSGVGPLNESKKDASAEDMEYHQNSSTLEVGH 63
H I+G +++ + + +ES +++ A + +YH +S + E H
Sbjct 39 HRQGIIGVEKMTNHIFSDSESDRESGASEFDYHSDSQSEETQH 81
> ath:AT1G29590 eukaryotic translation initiation factor 4E, putative
/ eIF-4E, putative / eIF4E, putative / mRNA cap-binding
protein, putative; K03259 translation initiation factor
4E
Length=285
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 31/67 (46%), Gaps = 12/67 (17%)
Query 8 DPGTMGGPCSHEYHVSSILGSDRIVSG--VGPLNESKKDASAEDMEYHQNSSTLEVGHCF 65
DP T P E HVS+I + +SG P E K AS ++++ ++ HCF
Sbjct 63 DPNTTTSPSPIEKHVSAI----KAISGDEKAPSKEKKNYAS------KKSTTVIQKSHCF 112
Query 66 QLPDTYC 72
Q T+
Sbjct 113 QNSWTFW 119
> pfa:PFF1595c VAR; erythrocyte membrane protein 1, PfEMP1; K13850
erythrocyte membrane protein 1
Length=2238
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 17/27 (62%), Gaps = 2/27 (7%)
Query 6 QADPGTMGGPC--SHEYHVSSILGSDR 30
++DP T G PC +EYH + LG D+
Sbjct 57 ESDPQTPGDPCQLQYEYHTNVTLGYDK 83
Lambda K H
0.320 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40