bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0814_orf1
Length=176
Score E
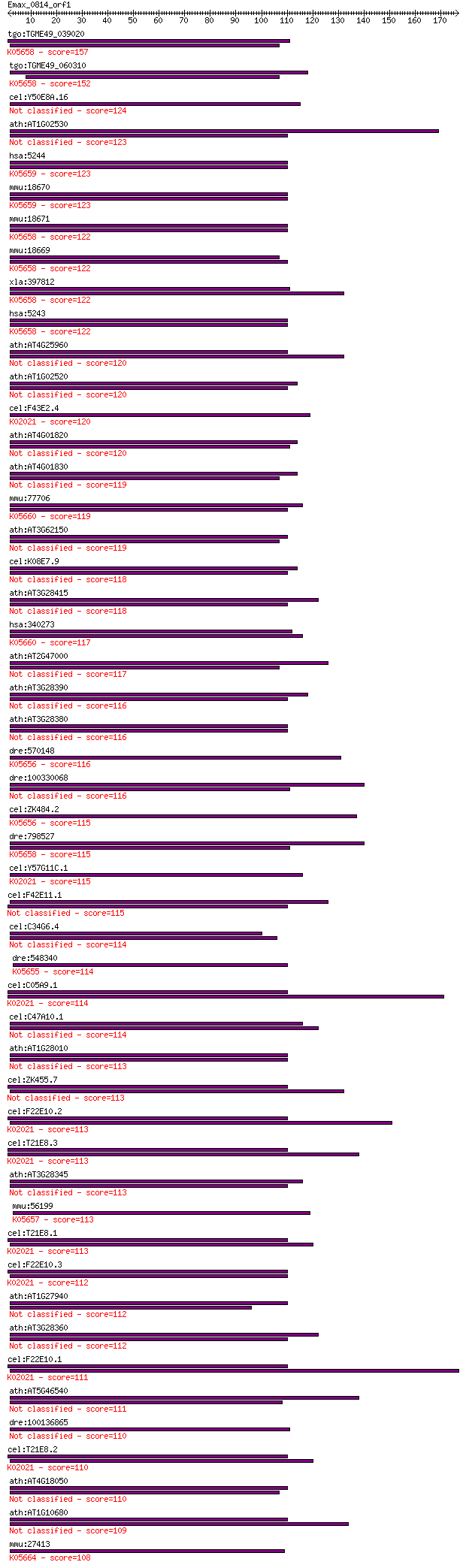
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_039020 ATP-binding cassette protein subfamily B mem... 157 2e-38
tgo:TGME49_060310 ATP-binding cassette protein subfamily B mem... 152 4e-37
cel:Y50E8A.16 haf-7; HAlF transporter (PGP related) family mem... 124 2e-28
ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, couple... 123 3e-28
hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;... 123 3e-28
mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassett... 123 4e-28
mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,... 122 5e-28
mmu:18669 Abcb1b, Abcb1, Mdr1, Mdr1b, Pgy-1, Pgy1, mdr; ATP-bi... 122 6e-28
xla:397812 abcb1, xemdr; ATP-binding cassette, sub-family B (M... 122 6e-28
hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296, P-... 122 6e-28
ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled t... 120 3e-27
ath:AT1G02520 PGP11; PGP11 (P-GLYCOPROTEIN 11); ATPase, couple... 120 3e-27
cel:F43E2.4 haf-2; HAlF transporter (PGP related) family membe... 120 3e-27
ath:AT4G01820 PGP3; PGP3 (P-GLYCOPROTEIN 3); ATPase, coupled t... 120 4e-27
ath:AT4G01830 PGP5; PGP5 (P-GLYCOPROTEIN 5); ATPase, coupled t... 119 6e-27
mmu:77706 Abcb5, 9230106F14Rik; ATP-binding cassette, sub-fami... 119 7e-27
ath:AT3G62150 PGP21; PGP21 (P-GLYCOPROTEIN 21); ATPase, couple... 119 8e-27
cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1) 118 1e-26
ath:AT3G28415 P-glycoprotein, putative 118 1e-26
hsa:340273 ABCB5, ABCB5alpha, ABCB5beta, EST422562; ATP-bindin... 117 2e-26
ath:AT2G47000 ABCB4; ABCB4 (ATP BINDING CASSETTE SUBFAMILY B4)... 117 2e-26
ath:AT3G28390 PGP18; PGP18 (P-GLYCOPROTEIN 18); ATPase, couple... 116 3e-26
ath:AT3G28380 PGP17; PGP17 (P-GLYCOPROTEIN 17); ATP binding / ... 116 3e-26
dre:570148 abcb9, si:dkey-21k4.3; ATP-binding cassette, sub-fa... 116 4e-26
dre:100330068 ATP-binding cassette, subfamily B, member 1B-like 116 5e-26
cel:ZK484.2 haf-9; HAlF transporter (PGP related) family membe... 115 6e-26
dre:798527 abcb5, im:7158730, mdr1, wu:fc18f02, wu:fi81f06; AT... 115 8e-26
cel:Y57G11C.1 haf-8; HAlF transporter (PGP related) family mem... 115 9e-26
cel:F42E11.1 pgp-4; P-GlycoProtein related family member (pgp-4) 115 1e-25
cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2) 114 1e-25
dre:548340 abcb8, zgc:113037; ATP-binding cassette, sub-family... 114 1e-25
cel:C05A9.1 pgp-5; P-GlycoProtein related family member (pgp-5... 114 2e-25
cel:C47A10.1 pgp-9; P-GlycoProtein related family member (pgp-9) 114 3e-25
ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, couple... 113 3e-25
cel:ZK455.7 pgp-3; P-GlycoProtein related family member (pgp-3) 113 3e-25
cel:F22E10.2 pgp-13; P-GlycoProtein related family member (pgp... 113 3e-25
cel:T21E8.3 pgp-8; P-GlycoProtein related family member (pgp-8... 113 3e-25
ath:AT3G28345 ABC transporter family protein 113 4e-25
mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-bindin... 113 4e-25
cel:T21E8.1 pgp-6; P-GlycoProtein related family member (pgp-6... 113 4e-25
cel:F22E10.3 pgp-14; P-GlycoProtein related family member (pgp... 112 6e-25
ath:AT1G27940 PGP13; PGP13 (P-GLYCOPROTEIN 13); ATPase, couple... 112 7e-25
ath:AT3G28360 PGP16; PGP16 (P-GLYCOPROTEIN 16); ATPase, couple... 112 8e-25
cel:F22E10.1 pgp-12; P-GlycoProtein related family member (pgp... 111 1e-24
ath:AT5G46540 PGP7; PGP7 (P-GLYCOPROTEIN 7); ATPase, coupled t... 111 2e-24
dre:100136865 abcb4, zgc:172149; ATP-binding cassette, sub-fam... 110 2e-24
cel:T21E8.2 pgp-7; P-GlycoProtein related family member (pgp-7... 110 3e-24
ath:AT4G18050 PGP9; PGP9 (P-GLYCOPROTEIN 9); ATPase, coupled t... 110 3e-24
ath:AT1G10680 PGP10; PGP10 (P-GLYCOPROTEIN 10); ATPase, couple... 109 4e-24
mmu:27413 Abcb11, ABC16, Bsep, Lith1, PFIC2, PGY4, SPGP; ATP-b... 108 7e-24
> tgo:TGME49_039020 ATP-binding cassette protein subfamily B member
2 (EC:3.6.3.30 3.6.3.44); K05658 ATP-binding cassette,
subfamily B (MDR/TAP), member 1
Length=1407
Score = 157 bits (396), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 79/110 (71%), Positives = 91/110 (82%), Gaps = 0/110 (0%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
GQLSGGQKQRI IARAL+R PSILIFDEATSALDT SE+VVQ+ALDSL+KTT ATTLIVA
Sbjct 583 GQLSGGQKQRIVIARALVRRPSILIFDEATSALDTVSEKVVQDALDSLIKTTNATTLIVA 642
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
HRL+TI+NAD I+VL+ G+ VVQ GTH+ LM GLYY +V SQ+
Sbjct 643 HRLTTIRNADQIIVLDNRDGTGSQVVQVGTHQSLMSQEDGLYYQLVQSQL 692
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 61/105 (58%), Positives = 80/105 (76%), Gaps = 0/105 (0%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+ P +LI DEATSALD +SER+VQ LD+++ T + TL++AH
Sbjct 1296 QLSGGQKQRIAIARALLTQPRMLILDEATSALDAESERIVQATLDNVIATKERVTLMIAH 1355
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RLST+++AD IVVL G+ VV+ GTH++LM G+Y H+V
Sbjct 1356 RLSTVRDADKIVVLSNEDKRGSQVVEVGTHDELMAIPDGVYRHLV 1400
> tgo:TGME49_060310 ATP-binding cassette protein subfamily B member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1345
Score = 152 bits (385), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 78/117 (66%), Positives = 93/117 (79%), Gaps = 1/117 (0%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARALIR PSILIFDEATSALD SERVVQ ALD L+++T TT+I+AH
Sbjct 530 QLSGGQKQRVAIARALIRHPSILIFDEATSALDNASERVVQAALDRLIESTGVTTIIIAH 589
Query 62 RLSTIQNADLIVVL-EPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGD 117
RLSTI+ ADLIVVL + G+ VVQQG+HE+LM+D GLY+ +V SQ+ GD
Sbjct 590 RLSTIRRADLIVVLGQRDDAGGSAVVQQGSHEELMKDESGLYFSLVQSQLAGLHGGD 646
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 75/100 (75%), Gaps = 1/100 (1%)
Query 8 KQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLV-KTTKATTLIVAHRLSTI 66
KQRIAIARAL+R P +LI DEATSALD +SER VQ+ LD L+ K K +T+IVAHRLST+
Sbjct 1239 KQRIAIARALVRKPRLLILDEATSALDPESERQVQKTLDELMEKGHKHSTIIVAHRLSTV 1298
Query 67 QNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
+NA+ IVVL G+ VV+ GTHE+LM+ G+Y +V
Sbjct 1299 RNANKIVVLSNEDGRGSRVVEVGTHEELMKLKNGVYRQLV 1338
> cel:Y50E8A.16 haf-7; HAlF transporter (PGP related) family member
(haf-7)
Length=807
Score = 124 bits (311), Expect = 2e-28, Method: Composition-based stats.
Identities = 63/113 (55%), Positives = 85/113 (75%), Gaps = 7/113 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARALIR P +LI DEATSALD++SE +VQEAL+ + + T L++AH
Sbjct 687 QMSGGQKQRIAIARALIRDPRVLILDEATSALDSESEAMVQEALNRCAR--ERTVLVIAH 744
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKI 114
RLST+++AD I V+E V + G HE+LM++T+GLYY +V+ Q+ P I
Sbjct 745 RLSTVRSADRIAVIEK-----GNVTEMGNHEELMKNTEGLYYKLVSKQLDPLI 792
> ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 66/167 (39%), Positives = 103/167 (61%), Gaps = 23/167 (13%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P +L+ DEATSALDT+SERVVQEALD ++ TT++VAH
Sbjct 506 QLSGGQKQRIAIARAILKDPRVLLLDEATSALDTESERVVQEALDRVM--VNRTTVVVAH 563
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLST++NAD+I V+ H +V++G+H +L++D+ G Y ++ Q + K
Sbjct 564 RLSTVRNADMIAVI-----HSGKMVEKGSHSELLKDSVGAYSQLIRCQEINK-------- 610
Query 122 AVQETIPAPHEPEPADGAAVPTLQERRMSTDFVAASIAKASRSLAKT 168
H+ +P+D A+ + + ++ + I+ + S +
Sbjct 611 --------GHDAKPSDMASGSSFRNSNLNISREGSVISGGTSSFGNS 649
Score = 112 bits (279), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 58/113 (51%), Positives = 80/113 (70%), Gaps = 12/113 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P IL+ DEATSALD +SER+VQ+ALD ++ TT++VAH
Sbjct 1168 QLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESERLVQDALDRVI--VNRTTVVVAH 1225
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLY-----YHMVASQ 109
RLSTI+NAD+I +++ + + GTHE L++ G+Y HM AS
Sbjct 1226 RLSTIKNADVIAIVKN-----GVIAENGTHETLIKIDGGVYASLVQLHMTASN 1273
> hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;
ATP-binding cassette, sub-family B (MDR/TAP), member 4 (EC:3.6.3.44);
K05659 ATP-binding cassette, subfamily B (MDR/TAP),
member 4
Length=1279
Score = 123 bits (308), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 67/108 (62%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARALIR P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1174 QLSGGQKQRIAIARALIRQPQILLLDEATSALDTESEKVVQEALDK--AREGRTCIVIAH 1231
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTIQNADLIVV + V + GTH+QL+ KG+Y+ MV+ Q
Sbjct 1232 RLSTIQNADLIVVFQ-----NGRVKEHGTHQQLLAQ-KGIYFSMVSVQ 1273
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 77/108 (71%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VQ ALD TT+++AH
Sbjct 532 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAEVQAALDK--AREGRTTIVIAH 589
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLST++NAD+I E +V+QG+H +LM+ +G+Y+ +V Q
Sbjct 590 RLSTVRNADVIAGFED-----GVIVEQGSHSELMKK-EGVYFKLVNMQ 631
> mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassette,
sub-family B (MDR/TAP), member 4 (EC:3.6.3.44); K05659
ATP-binding cassette, subfamily B (MDR/TAP), member 4
Length=1276
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 67/108 (62%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARALIR P +L+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1171 QLSGGQKQRIAIARALIRQPRVLLLDEATSALDTESEKVVQEALDK--AREGRTCIVIAH 1228
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTIQNADLIVV+E V + GTH+QL+ KG+Y+ MV Q
Sbjct 1229 RLSTIQNADLIVVIE-----NGKVKEHGTHQQLLAQ-KGIYFSMVNIQ 1270
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 77/108 (71%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VQ ALD TT+++AH
Sbjct 529 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAEVQAALDK--AREGRTTIVIAH 586
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+NAD+I E +V+QG+H +LM+ +G+Y+ +V Q
Sbjct 587 RLSTIRNADVIAGFED-----GVIVEQGSHSELMKK-EGIYFRLVNMQ 628
> mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,
Pgy3, mdr-3; ATP-binding cassette, sub-family B (MDR/TAP),
member 1A (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1276
Score = 122 bits (307), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 66/108 (61%), Positives = 81/108 (75%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1171 QLSGGQKQRIAIARALVRQPHILLLDEATSALDTESEKVVQEALDK--AREGRTCIVIAH 1228
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTIQNADLIVV++ V + GTH+QL+ KG+Y+ MV+ Q
Sbjct 1229 RLSTIQNADLIVVIQ-----NGKVKEHGTHQQLLAQ-KGIYFSMVSVQ 1270
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 62/108 (57%), Positives = 81/108 (75%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VVQ ALD TT+++AH
Sbjct 526 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAVVQAALDK--AREGRTTIVIAH 583
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLST++NAD+I + G +V+QG H++LMR+ KG+Y+ +V +Q
Sbjct 584 RLSTVRNADVI-----AGFDGGVIVEQGNHDELMRE-KGIYFKLVMTQ 625
> mmu:18669 Abcb1b, Abcb1, Mdr1, Mdr1b, Pgy-1, Pgy1, mdr; ATP-binding
cassette, sub-family B (MDR/TAP), member 1B (EC:3.6.3.44);
K05658 ATP-binding cassette, subfamily B (MDR/TAP), member
1
Length=1276
Score = 122 bits (306), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 66/105 (62%), Positives = 79/105 (75%), Gaps = 8/105 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1173 QLSGGQKQRIAIARALVRQPHILLLDEATSALDTESEKVVQEALDK--AREGRTCIVIAH 1230
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RLSTIQNADLIVV+E V + GTH+QL+ KG+Y+ MV
Sbjct 1231 RLSTIQNADLIVVIE-----NGKVKEHGTHQQLLAQ-KGIYFSMV 1269
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 62/108 (57%), Positives = 81/108 (75%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VVQ ALD TT+++AH
Sbjct 529 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAVVQAALDK--AREGRTTIVIAH 586
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLST++NAD+I + G +V+QG H++LMR+ KG+Y+ +V +Q
Sbjct 587 RLSTVRNADVI-----AGFDGGVIVEQGNHDELMRE-KGIYFKLVMTQ 628
> xla:397812 abcb1, xemdr; ATP-binding cassette, sub-family B
(MDR/TAP), member 1; K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1287
Score = 122 bits (306), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 66/109 (60%), Positives = 82/109 (75%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARALIR P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1184 QLSGGQKQRIAIARALIRKPKILLLDEATSALDTESEKVVQEALDK--ARMGRTCIVIAH 1241
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
RLSTIQNAD I V++ VV+QGTH+QL++ KG+Y+ +V Q+
Sbjct 1242 RLSTIQNADKIAVIQ-----NGKVVEQGTHQQLLQ-LKGVYFSLVTIQL 1284
Score = 112 bits (280), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 65/133 (48%), Positives = 87/133 (65%), Gaps = 11/133 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VVQ ALD TT++VAH
Sbjct 540 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAVVQSALDK--AREGRTTIVVAH 597
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDD- 120
RLSTI+NA+ I + +V+QG+H++LM + G+Y+++V Q V ++D
Sbjct 598 RLSTIRNANAI-----AGFDNGVIVEQGSHKELM-ERGGVYFNLVTLQTVETSKDTEEDL 651
Query 121 --SAVQETIPAPH 131
++ IP H
Sbjct 652 ETHIYEKKIPVTH 664
> hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296,
P-GP, PGY1; ATP-binding cassette, sub-family B (MDR/TAP), member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1280
Score = 122 bits (306), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 66/108 (61%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1175 QLSGGQKQRIAIARALVRQPHILLLDEATSALDTESEKVVQEALDK--AREGRTCIVIAH 1232
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTIQNADLIVV + V + GTH+QL+ KG+Y+ MV+ Q
Sbjct 1233 RLSTIQNADLIVVFQ-----NGRVKEHGTHQQLLAQ-KGIYFSMVSVQ 1274
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 80/108 (74%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE VVQ ALD K TT+++AH
Sbjct 530 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEAVVQVALDKARKGR--TTIVIAH 587
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLST++NAD+I + +V++G H++LM++ KG+Y+ +V Q
Sbjct 588 RLSTVRNADVIAGFDD-----GVIVEKGNHDELMKE-KGIYFKLVTMQ 629
> ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 120 bits (301), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 83/108 (76%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQ+QRIAIARA+++ P+IL+ DEATSALD +SERVVQ+ALD L+ TT++VAH
Sbjct 1168 QMSGGQRQRIAIARAILKNPAILLLDEATSALDVESERVVQQALDRLM--ANRTTVVVAH 1225
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+NAD I VL HG +V+QG+H +L+ + G Y+ +++ Q
Sbjct 1226 RLSTIKNADTISVL-----HGGKIVEQGSHRKLVLNKSGPYFKLISLQ 1268
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 60/130 (46%), Positives = 86/130 (66%), Gaps = 10/130 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAI+RA+++ PSIL+ DEATSALD +SE+ VQEALD ++ TT++VAH
Sbjct 539 QLSGGQKQRIAISRAIVKNPSILLLDEATSALDAESEKSVQEALDRVM--VGRTTVVVAH 596
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLST++NAD+I V+ H +V+ G HE L+ + G Y ++ Q + +
Sbjct 597 RLSTVRNADIIAVV-----HEGKIVEFGNHENLISNPDGAYSSLLRLQETASL---QRNP 648
Query 122 AVQETIPAPH 131
++ T+ PH
Sbjct 649 SLNRTLSRPH 658
> ath:AT1G02520 PGP11; PGP11 (P-GLYCOPROTEIN 11); ATPase, coupled
to transmembrane movement of substances
Length=1278
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 85/112 (75%), Gaps = 7/112 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SERVVQEALD ++ TT+IVAH
Sbjct 519 QLSGGQKQRIAIARAILKDPRILLLDEATSALDAESERVVQEALDRVM--VNRTTVIVAH 576
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK 113
RLST++NAD+I V+ H +V++G+H +L++D++G Y ++ Q + K
Sbjct 577 RLSTVRNADMIAVI-----HRGKMVEKGSHSELLKDSEGAYSQLIRLQEINK 623
Score = 114 bits (284), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/113 (53%), Positives = 81/113 (71%), Gaps = 12/113 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P IL+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1173 QLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESERVVQDALDRVM--VNRTTIVVAH 1230
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLY-----YHMVASQ 109
RLSTI+NAD+I V++ + ++GTHE L++ G+Y HM AS
Sbjct 1231 RLSTIKNADVIAVVK-----NGVIAEKGTHETLIKIEGGVYASLVQLHMTASN 1278
> cel:F43E2.4 haf-2; HAlF transporter (PGP related) family member
(haf-2); K02021 putative ABC transport system ATP-binding
protein
Length=761
Score = 120 bits (300), Expect = 3e-27, Method: Composition-based stats.
Identities = 63/117 (53%), Positives = 84/117 (71%), Gaps = 7/117 (5%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARAL+R P++LI DEATSALDT+SE +VQ+AL + + T +IVAH
Sbjct 652 QMSGGQKQRIAIARALVRNPAVLILDEATSALDTESEALVQQALSRCAQ--ERTVIIVAH 709
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDD 118
RLSTI+ A+ I V+ +VQ GTH +LM DT G+YY +V+ Q++ G+D
Sbjct 710 RLSTIEKANKIAVIVK-----GCLVQMGTHTELMTDTDGMYYSLVSRQMLSAKVGED 761
> ath:AT4G01820 PGP3; PGP3 (P-GLYCOPROTEIN 3); ATPase, coupled
to transmembrane movement of substances
Length=1229
Score = 120 bits (300), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 85/112 (75%), Gaps = 7/112 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SERVVQEALD ++ + TT+IVAH
Sbjct 486 QLSGGQKQRIAIARAILKDPRILLLDEATSALDAESERVVQEALDRVMMSR--TTVIVAH 543
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK 113
RLST++NAD+I V+ H +V++G+H +L++D +G Y ++ Q + K
Sbjct 544 RLSTVRNADMIAVI-----HRGKIVEEGSHSELLKDHEGAYAQLIRLQKIKK 590
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 59/109 (54%), Positives = 80/109 (73%), Gaps = 7/109 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P IL+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1124 QLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESERVVQDALDRVM--VNRTTVVVAH 1181
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
RLSTI+NAD+I V++ +V++GTHE L+ G+Y +V I
Sbjct 1182 RLSTIKNADVIAVVKN-----GVIVEKGTHETLINIEGGVYASLVQLHI 1225
> ath:AT4G01830 PGP5; PGP5 (P-GLYCOPROTEIN 5); ATPase, coupled
to transmembrane movement of substances
Length=1230
Score = 119 bits (298), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 84/112 (75%), Gaps = 7/112 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SERVVQEALD ++ TT+IVAH
Sbjct 491 QLSGGQKQRIAIARAILKDPRILLLDEATSALDAESERVVQEALDRIM--VNRTTVIVAH 548
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK 113
RLST++NAD+I V+ H +V++G+H +L++D +G Y ++ Q + K
Sbjct 549 RLSTVRNADIIAVI-----HRGKIVEEGSHSELLKDHEGAYSQLLRLQEINK 595
Score = 112 bits (279), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 57/105 (54%), Positives = 78/105 (74%), Gaps = 7/105 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P IL+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1125 QLSGGQKQRVAIARAIVKEPKILLLDEATSALDAESERVVQDALDRVM--VNRTTIVVAH 1182
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RLSTI+NAD+I V++ + ++GTHE L+ G+Y +V
Sbjct 1183 RLSTIKNADVIAVVKN-----GVIAEKGTHETLINIEGGVYASLV 1222
> mmu:77706 Abcb5, 9230106F14Rik; ATP-binding cassette, sub-family
B (MDR/TAP), member 5; K05660 ATP-binding cassette, subfamily
B (MDR/TAP), member 5
Length=1255
Score = 119 bits (297), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 66/115 (57%), Positives = 85/115 (73%), Gaps = 10/115 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKA-TTLIVA 60
Q+SGGQKQRIAIARAL+R P ILI DEATSALDT+SE +VQ AL+ K +K TT++VA
Sbjct 525 QMSGGQKQRIAIARALVRNPKILILDEATSALDTESESLVQTALE---KASKGRTTIVVA 581
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKID 115
HRLSTI+ ADLIV ++ VV++GTH +LM +GLYY + +Q + K+D
Sbjct 582 HRLSTIRGADLIVTMKD-----GMVVEKGTHAELMAK-QGLYYSLAMAQDIKKVD 630
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/108 (57%), Positives = 82/108 (75%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARAL+R P IL+ DEATSALD +SE+VVQ+ALD + T L+VAH
Sbjct 1156 QLSGGQKQRLAIARALLRKPKILLLDEATSALDNESEKVVQQALDKARR--GKTCLVVAH 1213
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTIQNAD+IVVL+ ++ +QGTH++L+R+ Y+ +VA+
Sbjct 1214 RLSTIQNADMIVVLQ-----NGSIKEQGTHQELLRNGD-TYFKLVAAH 1255
> ath:AT3G62150 PGP21; PGP21 (P-GLYCOPROTEIN 21); ATPase, coupled
to transmembrane movement of substances
Length=1292
Score = 119 bits (297), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 58/108 (53%), Positives = 82/108 (75%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIA+ARA+++ P IL+ DEATSALD +SER+VQEALD ++ TT++VAH
Sbjct 541 QLSGGQKQRIAVARAILKDPRILLLDEATSALDAESERIVQEALDRIM--VNRTTVVVAH 598
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLST++NAD+I V+ H +V++G+H +L+RD +G Y ++ Q
Sbjct 599 RLSTVRNADMIAVI-----HQGKIVEKGSHSELLRDPEGAYSQLIRLQ 641
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 78/105 (74%), Gaps = 7/105 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P +L+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1187 QLSGGQKQRVAIARAIVKDPKVLLLDEATSALDAESERVVQDALDRVM--VNRTTVVVAH 1244
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RLSTI+NAD+I V++ +V++G HE L+ G+Y +V
Sbjct 1245 RLSTIKNADVIAVVK-----NGVIVEKGKHETLINIKDGVYASLV 1284
> cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1)
Length=1321
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 62/112 (55%), Positives = 79/112 (70%), Gaps = 8/112 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALDT+SE+VVQEALD T +++AH
Sbjct 1217 QLSGGQKQRIAIARALVRNPKILLLDEATSALDTESEKVVQEALDR--AREGRTCIVIAH 1274
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK 113
RL+T+ NAD I V+ T++++GTH QLM + KG YY + Q+ K
Sbjct 1275 RLNTVMNADCIAVVS-----NGTIIEKGTHTQLMSE-KGAYYKLTQKQMTEK 1320
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 65/108 (60%), Positives = 79/108 (73%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P IL+ DEATSALD +SE +VQ+ALD K TT+I+AH
Sbjct 554 QLSGGQKQRIAIARALVRNPKILLLDEATSALDAESEGIVQQALDKAAK--GRTTIIIAH 611
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+NADLI+ S +G VV+ G H LM +GLYY +V +Q
Sbjct 612 RLSTIRNADLII----SCKNG-QVVEVGDHRALMAQ-QGLYYDLVTAQ 653
> ath:AT3G28415 P-glycoprotein, putative
Length=1221
Score = 118 bits (295), Expect = 1e-26, Method: Composition-based stats.
Identities = 61/120 (50%), Positives = 87/120 (72%), Gaps = 7/120 (5%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRI+IARA+I+ P++L+ DEATSALD++SERVVQEALD+ T TT+++AH
Sbjct 476 QMSGGQKQRISIARAIIKSPTLLLLDEATSALDSESERVVQEALDN--ATIGRTTIVIAH 533
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLSTI+N D+I V + +V+ G+HE+LM + G Y +V QI+ + +D+ S
Sbjct 534 RLSTIRNVDVICVFK-----NGQIVETGSHEELMENVDGQYTSLVRLQIMENEESNDNVS 588
Score = 107 bits (267), Expect = 2e-23, Method: Composition-based stats.
Identities = 59/109 (54%), Positives = 79/109 (72%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ PS+L+ DEATSALD QSER+VQ+AL L+ T++++AH
Sbjct 1116 QLSGGQKQRIAIARAVLKNPSVLLLDEATSALDNQSERMVQDALGRLM--VGRTSVVIAH 1173
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM-RDTKGLYYHMVASQ 109
RLSTIQN D I VL+ VV+ GTH L+ + G+Y+ +V+ Q
Sbjct 1174 RLSTIQNCDTITVLDK-----GKVVECGTHSSLLAKGPTGVYFSLVSLQ 1217
> hsa:340273 ABCB5, ABCB5alpha, ABCB5beta, EST422562; ATP-binding
cassette, sub-family B (MDR/TAP), member 5; K05660 ATP-binding
cassette, subfamily B (MDR/TAP), member 5
Length=1257
Score = 117 bits (294), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 63/110 (57%), Positives = 80/110 (72%), Gaps = 8/110 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARAL++ P IL+ DEATSALD SE+VVQ ALD T T L+V H
Sbjct 1155 QLSGGQKQRLAIARALLQKPKILLLDEATSALDNDSEKVVQHALDK--ARTGRTCLVVTH 1212
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIV 111
RLS IQNADLIVVL H + +QGTH++L+R+ + +Y+ +V +Q V
Sbjct 1213 RLSAIQNADLIVVL-----HNGKIKEQGTHQELLRN-RDIYFKLVNAQSV 1256
Score = 112 bits (281), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 64/115 (55%), Positives = 83/115 (72%), Gaps = 10/115 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKA-TTLIVA 60
Q+SGGQKQRIAIARAL+R P ILI DEATSALD++S+ VQ AL+ K +K TT++VA
Sbjct 524 QMSGGQKQRIAIARALVRNPKILILDEATSALDSESKSAVQAALE---KASKGRTTIVVA 580
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKID 115
HRLSTI++ADLIV L+ + ++G H +LM +GLYY +V SQ + K D
Sbjct 581 HRLSTIRSADLIVTLKD-----GMLAEKGAHAELMAK-RGLYYSLVMSQDIKKAD 629
> ath:AT2G47000 ABCB4; ABCB4 (ATP BINDING CASSETTE SUBFAMILY B4);
ATPase, coupled to transmembrane movement of substances
/ xenobiotic-transporting ATPase
Length=1286
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 62/124 (50%), Positives = 89/124 (71%), Gaps = 11/124 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIA+ARA+++ P IL+ DEATSALD +SERVVQEALD ++ TT++VAH
Sbjct 522 QLSGGQKQRIAVARAILKDPRILLLDEATSALDAESERVVQEALDRIM--VNRTTVVVAH 579
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLST++NAD+I V+ H +V++G+H +L++D +G Y ++ Q K D++
Sbjct 580 RLSTVRNADMIAVI-----HQGKIVEKGSHTELLKDPEGAYSQLIRLQEEKK----SDEN 630
Query 122 AVQE 125
A +E
Sbjct 631 AAEE 634
Score = 108 bits (271), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 55/105 (52%), Positives = 78/105 (74%), Gaps = 7/105 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ P +L+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1181 QLSGGQKQRVAIARAIVKDPKVLLLDEATSALDAESERVVQDALDRVM--VNRTTIVVAH 1238
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RLSTI+NAD+I V++ +V++G H+ L+ G+Y +V
Sbjct 1239 RLSTIKNADVIAVVK-----NGVIVEKGKHDTLINIKDGVYASLV 1278
> ath:AT3G28390 PGP18; PGP18 (P-GLYCOPROTEIN 18); ATPase, coupled
to transmembrane movement of substances
Length=1225
Score = 116 bits (291), Expect = 3e-26, Method: Composition-based stats.
Identities = 62/116 (53%), Positives = 83/116 (71%), Gaps = 7/116 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+I+ P IL+ DEATSALD++SERVVQEALD+ + TT+++AH
Sbjct 485 QLSGGQKQRIAIARAIIKSPIILLLDEATSALDSESERVVQEALDN--ASIGRTTIVIAH 542
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGD 117
RLSTI+NAD+I V+ H +++ G+HE+L+ G Y +V Q V + D
Sbjct 543 RLSTIRNADVICVV-----HNGRIIETGSHEELLEKLDGQYTSLVRLQQVDNKESD 593
Score = 107 bits (267), Expect = 2e-23, Method: Composition-based stats.
Identities = 60/109 (55%), Positives = 79/109 (72%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ PS+L+ DEATSALD+QSE VVQ+AL+ L+ T++++AH
Sbjct 1120 QLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSESVVQDALERLM--VGRTSVVIAH 1177
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM-RDTKGLYYHMVASQ 109
RLSTIQ D I VLE A VV+ G H L+ + KG Y+ +V+ Q
Sbjct 1178 RLSTIQKCDTIAVLENGA-----VVECGNHSSLLAKGPKGAYFSLVSLQ 1221
> ath:AT3G28380 PGP17; PGP17 (P-GLYCOPROTEIN 17); ATP binding
/ ATPase/ ATPase, coupled to transmembrane movement of substances
/ nucleoside-triphosphatase/ nucleotide binding
Length=1240
Score = 116 bits (291), Expect = 3e-26, Method: Composition-based stats.
Identities = 59/108 (54%), Positives = 82/108 (75%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARA+I+ P IL+ DEATSALD++SERVVQE+LD+ + TT+++AH
Sbjct 497 QMSGGQKQRIAIARAIIKSPKILLLDEATSALDSESERVVQESLDN--ASIGRTTIVIAH 554
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+NAD+I V+ H +V+ G+HE+L++ G Y +V+ Q
Sbjct 555 RLSTIRNADVICVI-----HNGQIVETGSHEELLKRIDGQYTSLVSLQ 597
Score = 106 bits (264), Expect = 4e-23, Method: Composition-based stats.
Identities = 57/109 (52%), Positives = 79/109 (72%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ PS+L+ DEATSALD++SERVVQ+AL+ ++ T++++AH
Sbjct 1135 QLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSKSERVVQDALERVM--VGRTSIMIAH 1192
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM-RDTKGLYYHMVASQ 109
RLSTIQN D+IVVL +V+ GTH L+ + G Y+ + Q
Sbjct 1193 RLSTIQNCDMIVVLGK-----GKIVESGTHSSLLEKGPTGTYFSLAGIQ 1236
> dre:570148 abcb9, si:dkey-21k4.3; ATP-binding cassette, sub-family
B (MDR/TAP), member 9; K05656 ATP-binding cassette, subfamily
B (MDR/TAP), member 9
Length=789
Score = 116 bits (291), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 93/130 (71%), Gaps = 14/130 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARALIR P ILI DEATSALD++SE +VQ+AL++L++ + T L++AH
Sbjct 649 QLSGGQKQRVAIARALIRSPRILILDEATSALDSESEYIVQQALNNLMR--EHTVLVIAH 706
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIV-PKIDGDDDD 120
RLST++ AD I+V++ +VV+QG H +LM GLY +V QI+ +ID DD +
Sbjct 707 RLSTVERADNILVIDK-----GSVVEQGPHAELMA-RGGLYCKLVQRQILGTEIDVDDKN 760
Query 121 SAVQETIPAP 130
+ PAP
Sbjct 761 PS-----PAP 765
> dre:100330068 ATP-binding cassette, subfamily B, member 1B-like
Length=1063
Score = 116 bits (290), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 69/145 (47%), Positives = 94/145 (64%), Gaps = 15/145 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL++ P IL+ DEATSALDTQSE +VQ ALD TT+++AH
Sbjct 301 QLSGGQKQRIAIARALVKNPKILLLDEATSALDTQSESIVQAALDK--ARAGRTTIVIAH 358
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK----IDGD 117
RLSTI++AD+I + VV+QG+H +LM KG+YY +V Q + +D +
Sbjct 359 RLSTIRSADII-----AGFSEGRVVEQGSHRELMAK-KGVYYSLVTQQTSGRQNEELDAN 412
Query 118 DDDS---AVQETIPAPHEPEPADGA 139
+DD+ + +ET +PE +G
Sbjct 413 EDDTQDDSEEETGEDSSDPEILEGG 437
Score = 115 bits (288), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 80/109 (73%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P +L+ DEATSALDT+SE++VQ ALD T +++AH
Sbjct 960 QLSGGQKQRIAIARALVRKPKLLLLDEATSALDTESEKIVQAALDE--ARLGRTCIVIAH 1017
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
RL+TIQNAD+IVV++ VV+QGTH QLM + Y+ +V +Q+
Sbjct 1018 RLTTIQNADIIVVVQ-----NGKVVEQGTHAQLMAKQEA-YFALVNAQV 1060
> cel:ZK484.2 haf-9; HAlF transporter (PGP related) family member
(haf-9); K05656 ATP-binding cassette, subfamily B (MDR/TAP),
member 9
Length=815
Score = 115 bits (289), Expect = 6e-26, Method: Composition-based stats.
Identities = 66/135 (48%), Positives = 87/135 (64%), Gaps = 17/135 (12%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARAL+R P +L+ DEATSALD +SE VQEA+ +K T +++AH
Sbjct 661 QMSGGQKQRIAIARALVRQPVVLLLDEATSALDAESEHTVQEAISKNLKGK--TVILIAH 718
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLST++NAD IVV+ + V Q G H+ LM + +GLY +V Q++ DG DD+
Sbjct 719 RLSTVENADKIVVI-----NKGKVEQLGNHKTLM-EQEGLYKQLVQRQMMSGEDGLDDEI 772
Query 122 AVQETIPAPHEPEPA 136
EPEPA
Sbjct 773 ---------EEPEPA 778
> dre:798527 abcb5, im:7158730, mdr1, wu:fc18f02, wu:fi81f06;
ATP-binding cassette, sub-family B (MDR/TAP), member 5; K05658
ATP-binding cassette, subfamily B (MDR/TAP), member 1
Length=1340
Score = 115 bits (288), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 69/145 (47%), Positives = 94/145 (64%), Gaps = 15/145 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL++ P IL+ DEATSALDTQSE +VQ ALD TT+++AH
Sbjct 576 QLSGGQKQRIAIARALVKNPKILLLDEATSALDTQSESIVQAALDK--ARAGRTTIVIAH 633
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPK----IDGD 117
RLSTI++AD+I + VV+QG+H +LM KG+YY +V Q + +D +
Sbjct 634 RLSTIRSADII-----AGFSEGRVVEQGSHRELMAK-KGVYYSLVTQQTSGRQNEELDAN 687
Query 118 DDDS---AVQETIPAPHEPEPADGA 139
+DD+ + +ET +PE +G
Sbjct 688 EDDTQDDSEEETGEDSSDPEILEGG 712
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 80/109 (73%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R P +L+ DEATSALDT+SE++VQ ALD T +++AH
Sbjct 1237 QLSGGQKQRIAIARALVRKPKLLLLDEATSALDTESEKIVQAALDE--ARLGRTCIVIAH 1294
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
RL+TIQNAD+IVV++ VV+QGTH QLM + Y+ +V +Q+
Sbjct 1295 RLTTIQNADIIVVVQ-----NGKVVEQGTHAQLMAKQEA-YFALVNAQV 1337
> cel:Y57G11C.1 haf-8; HAlF transporter (PGP related) family member
(haf-8); K02021 putative ABC transport system ATP-binding
protein
Length=633
Score = 115 bits (287), Expect = 9e-26, Method: Composition-based stats.
Identities = 59/114 (51%), Positives = 81/114 (71%), Gaps = 7/114 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARALIR P +LI DEATSALD++SE +VQEAL+ K + T +++AH
Sbjct 511 QMSGGQKQRIAIARALIRNPRVLILDEATSALDSESEGMVQEALNKCAK--ERTVIVIAH 568
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKID 115
RLST++ A I V+E +V + G H++LM + GLYY +V+ Q+ +D
Sbjct 569 RLSTVRKAQKIAVIE-----NGSVKEMGCHDELMENQDGLYYKLVSKQLGSIVD 617
> cel:F42E11.1 pgp-4; P-GlycoProtein related family member (pgp-4)
Length=1266
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 66/127 (51%), Positives = 87/127 (68%), Gaps = 11/127 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA++R P IL+ DEATSALDT+SER+VQ ALD + TTL +AH
Sbjct 512 QLSGGQKQRVAIARAIVRKPQILLLDEATSALDTESERMVQTALDK--ASEGRTTLCIAH 569
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGD---D 118
RLSTI+NA I+V + + ++G H+QL+R G+Y +MV +Q + K D D
Sbjct 570 RLSTIRNASKILVFDQ-----GLIPERGIHDQLIRQ-NGIYANMVRAQEIEKAKDDTTQD 623
Query 119 DDSAVQE 125
DD V+E
Sbjct 624 DDELVEE 630
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 60/109 (55%), Positives = 78/109 (71%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
G+LSGGQKQRIAIARA++R P IL+ DEATSALDT+SE++VQEALD T +++A
Sbjct 1164 GRLSGGQKQRIAIARAIVRNPKILLLDEATSALDTESEKIVQEALDK--ARLGRTCVVIA 1221
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLSTIQNAD I+V +++GTH+ L+ +GLYY +V Q
Sbjct 1222 HRLSTIQNADKIIV-----CRNGKAIEEGTHQTLLA-RRGLYYRLVEKQ 1264
> cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2)
Length=1272
Score = 114 bits (286), Expect = 1e-25, Method: Composition-based stats.
Identities = 56/98 (57%), Positives = 77/98 (78%), Gaps = 7/98 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL+R PS+L+ DEATSALDT+SE++VQEALD+ T L++AH
Sbjct 1167 QLSGGQKQRIAIARALVRSPSVLLLDEATSALDTESEKIVQEALDA--AKQGRTCLVIAH 1224
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTK 99
RLSTIQN+D+I ++ +V++GTH++L+R ++
Sbjct 1225 RLSTIQNSDVIAIVSE-----GKIVEKGTHDELIRKSE 1257
Score = 109 bits (273), Expect = 5e-24, Method: Composition-based stats.
Identities = 59/104 (56%), Positives = 74/104 (71%), Gaps = 8/104 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARAL++ P IL+ DEATSALDT++ER VQ ALD TT+IVAH
Sbjct 537 QLSGGQKQRIAIARALVKNPKILLLDEATSALDTEAEREVQGALDQ--AQAGRTTIIVAH 594
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHM 105
RLSTI+N D I V + +V+ G+HE+LM +G++Y M
Sbjct 595 RLSTIRNVDRIFVFK-----AGNIVESGSHEELM-SKQGIFYDM 632
> dre:548340 abcb8, zgc:113037; ATP-binding cassette, sub-family
B (MDR/TAP), member 8; K05655 ATP-binding cassette, subfamily
B (MDR/TAP), member 8
Length=714
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 65/107 (60%), Positives = 76/107 (71%), Gaps = 8/107 (7%)
Query 3 LSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAHR 62
LSGGQKQRIAIARAL++ PSILI DEATSALD +SERVVQEALD TT T LI+AHR
Sbjct 610 LSGGQKQRIAIARALVKNPSILILDEATSALDAESERVVQEALDR--ATTGRTVLIIAHR 667
Query 63 LSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
LSTIQ ADLI V+ +V+ GTH +L+ GLY ++ Q
Sbjct 668 LSTIQAADLICVMS-----NGRIVEAGTHLELL-SKGGLYAELIKRQ 708
> cel:C05A9.1 pgp-5; P-GlycoProtein related family member (pgp-5);
K02021 putative ABC transport system ATP-binding protein
Length=1252
Score = 114 bits (285), Expect = 2e-25, Method: Composition-based stats.
Identities = 59/109 (54%), Positives = 80/109 (73%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAIARA++R P +L+ DEATSALD+ SE+VVQ ALD+ + + +T++VA
Sbjct 1143 AQLSGGQKQRIAIARAILRNPKVLLLDEATSALDSDSEKVVQNALDT--ASERLSTVVVA 1200
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLST+ NAD I VL+ V +QGTHE+L+R + +Y+ +V Q
Sbjct 1201 HRLSTVVNADSIAVLK-----NGKVAEQGTHEELLR-KRSIYWRLVQKQ 1243
Score = 108 bits (270), Expect = 9e-24, Method: Composition-based stats.
Identities = 69/170 (40%), Positives = 101/170 (59%), Gaps = 11/170 (6%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAIAR L+R P IL+ DEATSALD +SE +VQEAL + TT++VA
Sbjct 511 AQLSGGQKQRIAIARTLVRNPRILLLDEATSALDNESEFIVQEALQK--ASIGRTTIVVA 568
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDD 120
HRLSTI+NA+ I+V+E +V+ G H+QL+ G+Y ++V +Q++ ++
Sbjct 569 HRLSTIRNANKIIVMEK-----GEIVEVGDHKQLIA-MNGVYNNLVQTQLMSTNYEKMNE 622
Query 121 SAVQETIPAPHEPEPADGAAVPTLQERRMSTDFVAASIAKASRSLAKTPN 170
+ + T + H P++ + Q+ D+V IA+ AK N
Sbjct 623 NEERVTRQSSHSDFPSNEI---SHQKIDQEDDYVKKLIAEIKEEGAKKSN 669
> cel:C47A10.1 pgp-9; P-GlycoProtein related family member (pgp-9)
Length=1294
Score = 114 bits (284), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 59/114 (51%), Positives = 84/114 (73%), Gaps = 8/114 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARAL+R P IL+ DEATSALD +SE +VQ AL++ + TT+++AH
Sbjct 521 QMSGGQKQRIAIARALVRNPKILLLDEATSALDAESESIVQSALEN--ASRGRTTIVIAH 578
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKID 115
RLST++NAD I+V++ V++ GTHE L+ + KGLY+ +V +Q+ +D
Sbjct 579 RLSTVRNADKIIVMK-----AGQVMEVGTHETLI-EQKGLYHELVHAQVFADVD 626
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 64/120 (53%), Positives = 80/120 (66%), Gaps = 8/120 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARALIR P IL+ DEATSALDT+SE+ VQ ALD+ K T ++VAH
Sbjct 1170 QLSGGQKQRIAIARALIRNPKILLLDEATSALDTESEKQVQVALDAAAKDR--TCIVVAH 1227
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLSTI NA I+V++ VV+QGTH +L+ +G Y+ + Q + G D S
Sbjct 1228 RLSTIVNAGCIMVVK-----NGQVVEQGTHNELIAK-RGAYFALTQKQSSNQSGGAFDTS 1281
> ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, coupled
to transmembrane movement of substances
Length=1247
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/108 (55%), Positives = 80/108 (74%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ PS+L+ DEATSALDT +E+ VQEALD L+K TT++VAH
Sbjct 1144 QLSGGQKQRVAIARAVLKDPSVLLLDEATSALDTSAEKQVQEALDKLMKGR--TTILVAH 1201
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+ AD IVVL H VV++G+H +L+ + G Y + + Q
Sbjct 1202 RLSTIRKADTIVVL-----HKGKVVEKGSHRELVSKSDGFYKKLTSLQ 1244
Score = 103 bits (258), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 58/108 (53%), Positives = 78/108 (72%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA++R P IL+ DEATSALD +SE++VQ+ALD++++ K TT+++AH
Sbjct 510 QLSGGQKQRIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVME--KRTTIVIAH 567
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+N D IVVL V + G+H +L+ G Y +V Q
Sbjct 568 RLSTIRNVDKIVVLRD-----GQVRETGSHSELI-SRGGDYATLVNCQ 609
> cel:ZK455.7 pgp-3; P-GlycoProtein related family member (pgp-3)
Length=1268
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 59/109 (54%), Positives = 78/109 (71%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
G+LSGGQKQR+AIARA++R P IL+ DEATSALDT+SE++VQEALD T +++A
Sbjct 1166 GRLSGGQKQRVAIARAIVRDPKILLLDEATSALDTESEKIVQEALDK--ARLGRTCVVIA 1223
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLSTIQNAD I+V +++GTH+ L+ +GLYY +V Q
Sbjct 1224 HRLSTIQNADKIIV-----CRNGKAIEEGTHQTLLA-RRGLYYRLVEKQ 1266
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 63/132 (47%), Positives = 87/132 (65%), Gaps = 9/132 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA++R P IL+ DEATSALDT+SER+VQ ALD + TTL +AH
Sbjct 514 QLSGGQKQRVAIARAIVRKPQILLLDEATSALDTESERMVQTALDK--ASEGRTTLCIAH 571
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDD--D 119
RLSTI+NA I+V + + ++GTH++L+ G+Y MV +Q + + D D
Sbjct 572 RLSTIRNASKILVFDQ-----GLIAERGTHDELISKDDGIYASMVKAQEIERAKEDTTLD 626
Query 120 DSAVQETIPAPH 131
D ++T + H
Sbjct 627 DEEDEKTHRSFH 638
> cel:F22E10.2 pgp-13; P-GlycoProtein related family member (pgp-13);
K02021 putative ABC transport system ATP-binding protein
Length=1324
Score = 113 bits (283), Expect = 3e-25, Method: Composition-based stats.
Identities = 61/109 (55%), Positives = 78/109 (71%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
GQLSGGQKQRIAIARAL+R P IL+ DEATSALD++SER VQEALD T + +A
Sbjct 1220 GQLSGGQKQRIAIARALVRDPKILLLDEATSALDSESERAVQEALDR--AREGRTCITIA 1277
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLS+IQN+DLIV ++ V + GTH++LM+ KG Y+ ++ Q
Sbjct 1278 HRLSSIQNSDLIVYID-----DGRVQESGTHKELMQ-LKGKYFELIKKQ 1320
Score = 108 bits (270), Expect = 1e-23, Method: Composition-based stats.
Identities = 67/149 (44%), Positives = 93/149 (62%), Gaps = 16/149 (10%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIAR LIR P +L+ DEATSALD QSE VVQ AL++ K TT+++AH
Sbjct 555 QLSGGQKQRVAIARTLIRDPKVLLLDEATSALDAQSESVVQSALNNASKGR--TTIMIAH 612
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLSTI+ AD IV E +V+ G HE+L+ + G Y+ +V +Q + D D+
Sbjct 613 RLSTIREADKIVFFEK-----GVIVEAGNHEELV-NLGGRYFDLVKAQAFKQ---DPDEI 663
Query 122 AVQETIPAPHEPEPADGAAVPTLQERRMS 150
A+++ E + D PT+ R++S
Sbjct 664 ALEK-----EEEDQFDEFDKPTVFNRKVS 687
> cel:T21E8.3 pgp-8; P-GlycoProtein related family member (pgp-8);
K02021 putative ABC transport system ATP-binding protein
Length=1243
Score = 113 bits (282), Expect = 3e-25, Method: Composition-based stats.
Identities = 61/109 (55%), Positives = 78/109 (71%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAI RA++R P +L+ DEATSALDT+SE++VQ ALD+ + + +T++VA
Sbjct 1140 AQLSGGQKQRIAITRAILRNPKLLLLDEATSALDTESEKIVQNALDT--ASERLSTIVVA 1197
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLSTI NAD I VL VV+QGTH QL+ KG Y+ +V Q
Sbjct 1198 HRLSTIINADSIAVLR-----NGKVVEQGTHNQLLA-VKGDYWRLVQHQ 1240
Score = 101 bits (251), Expect = 1e-21, Method: Composition-based stats.
Identities = 60/143 (41%), Positives = 89/143 (62%), Gaps = 14/143 (9%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAIAR L++ P+IL+ DEATSALD+ SER VQ AL + TT+I+A
Sbjct 517 AQLSGGQKQRIAIARVLVKNPAILLLDEATSALDSASERAVQLALKK--ASEGRTTIIIA 574
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ------IVPKI 114
HRLSTI++ D I+V+ + + G+HE+L+ + Y ++V +Q + I
Sbjct 575 HRLSTIRHCDKIMVMS-----NGKIAEVGSHEELISMDRE-YSNLVRAQFFDSQSVEEDI 628
Query 115 DGDDDDSAVQETIPAPHEPEPAD 137
+G + +Q+T P ++ EP +
Sbjct 629 NGQGAEEVIQKTPPNLNDGEPLE 651
> ath:AT3G28345 ABC transporter family protein
Length=1240
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 60/114 (52%), Positives = 83/114 (72%), Gaps = 7/114 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARA+I+ P+IL+ DEATSALD++SERVVQEAL++ + TT+++AH
Sbjct 497 QMSGGQKQRIAIARAIIKSPTILLLDEATSALDSESERVVQEALEN--ASIGRTTILIAH 554
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKID 115
RLSTI+NAD+I S +V+ G+H++LM + G Y +V Q + K D
Sbjct 555 RLSTIRNADVI-----SVVKNGHIVETGSHDELMENIDGQYSTLVHLQQIEKQD 603
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/109 (53%), Positives = 82/109 (75%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ PS+L+ DEATSALD+QSERVVQ+AL+ ++ T++++AH
Sbjct 1135 QLSGGQKQRIAIARAVLKNPSVLLLDEATSALDSQSERVVQDALERVM--VGRTSVVIAH 1192
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM-RDTKGLYYHMVASQ 109
RLSTIQN D I VL+ +V++GTH L+ + G+Y+ +V+ Q
Sbjct 1193 RLSTIQNCDAIAVLDK-----GKLVERGTHSSLLSKGPTGIYFSLVSLQ 1236
> mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-binding
cassette, sub-family B (MDR/TAP), member 10; K05657 ATP-binding
cassette, subfamily B (MDR/TAP), member 10
Length=715
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 60/116 (51%), Positives = 81/116 (69%), Gaps = 7/116 (6%)
Query 3 LSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAHR 62
LSGGQKQRIAIARAL++ P IL+ DEATSALD ++E +VQEALD L++ T LI+AHR
Sbjct 599 LSGGQKQRIAIARALLKNPKILLLDEATSALDAENEHLVQEALDRLMEGR--TVLIIAHR 656
Query 63 LSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDD 118
LSTI+NA+ + VL+ HG + + GTHE+L+ GLY ++ Q +G +
Sbjct 657 LSTIKNANFVAVLD----HG-KICEHGTHEELLLKPNGLYRKLMNKQSFLSYNGAE 707
> cel:T21E8.1 pgp-6; P-GlycoProtein related family member (pgp-6);
K02021 putative ABC transport system ATP-binding protein
Length=1263
Score = 113 bits (282), Expect = 4e-25, Method: Composition-based stats.
Identities = 59/109 (54%), Positives = 80/109 (73%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAIARA++R P +L+ DEATSALD+ SE+VVQ ALD+ + + +T++VA
Sbjct 1154 AQLSGGQKQRIAIARAILRNPKVLLLDEATSALDSDSEKVVQNALDT--ASERLSTVVVA 1211
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLST+ NAD I VL+ V +QGTHE+L+R + +Y+ +V Q
Sbjct 1212 HRLSTVVNADSIAVLK-----NGKVAEQGTHEELLR-KRSIYWRLVQKQ 1254
Score = 105 bits (262), Expect = 8e-23, Method: Composition-based stats.
Identities = 57/118 (48%), Positives = 84/118 (71%), Gaps = 8/118 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIAR L+R P IL+ DEATSALD +SE+VVQ+AL++ + TT++VAH
Sbjct 519 QLSGGQKQRIAIARTLVRNPKILLLDEATSALDNESEQVVQKALEN--ASQGRTTIVVAH 576
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDD 119
RLSTI+NA I+V++ +V+ G H++L+ +G+Y +V +Q++ D ++
Sbjct 577 RLSTIRNASKIIVMQK-----GEIVEVGNHDELIA-KRGVYNDLVQAQLLESHDDHEE 628
> cel:F22E10.3 pgp-14; P-GlycoProtein related family member (pgp-14);
K02021 putative ABC transport system ATP-binding protein
Length=1327
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 62/109 (56%), Positives = 75/109 (68%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
GQLSGGQKQRIAIARAL+R P IL+ DEATSALD++SER VQEALD T + +A
Sbjct 1223 GQLSGGQKQRIAIARALVRDPKILLLDEATSALDSESERAVQEALDR--AREGRTCITIA 1280
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLS+IQN+DLIV ++ V + G H QLM KG YY ++ Q
Sbjct 1281 HRLSSIQNSDLIVYIDK-----GKVQEAGNHTQLMHQ-KGRYYKLIKKQ 1323
Score = 106 bits (264), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 58/108 (53%), Positives = 75/108 (69%), Gaps = 8/108 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIAR LIR P +L+ DEATSALD QSE +VQ AL++ K TT+++AH
Sbjct 567 QLSGGQKQRVAIARTLIRDPKVLLLDEATSALDAQSESIVQSALNNASKGR--TTIMIAH 624
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+ AD IV E +V+ G HE+L+R G Y+ +V +Q
Sbjct 625 RLSTIREADKIVFFEK-----GVIVEAGNHEELVR-LGGRYFDLVKAQ 666
> ath:AT1G27940 PGP13; PGP13 (P-GLYCOPROTEIN 13); ATPase, coupled
to transmembrane movement of substances
Length=1245
Score = 112 bits (279), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 59/108 (54%), Positives = 79/108 (73%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIARA+++ PS+L+ DEATSALDT SE++VQEALD L+K TT++VAH
Sbjct 1142 QLSGGQKQRVAIARAVLKDPSVLLLDEATSALDTSSEKLVQEALDKLMKGR--TTVLVAH 1199
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+ AD + VL H VV++G+H +L+ G Y + + Q
Sbjct 1200 RLSTIRKADTVAVL-----HKGRVVEKGSHRELVSIPNGFYKQLTSLQ 1242
Score = 106 bits (264), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 56/94 (59%), Positives = 73/94 (77%), Gaps = 7/94 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA++R P IL+ DEATSALD +SE++VQ+ALD++++ K TT++VAH
Sbjct 509 QLSGGQKQRIAIARAVLRNPKILLLDEATSALDAESEKIVQQALDNVME--KRTTIVVAH 566
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM 95
RLSTI+N D IVVL V + G+H +LM
Sbjct 567 RLSTIRNVDKIVVLRD-----GQVRETGSHSELM 595
> ath:AT3G28360 PGP16; PGP16 (P-GLYCOPROTEIN 16); ATPase, coupled
to transmembrane movement of substances
Length=1228
Score = 112 bits (279), Expect = 8e-25, Method: Composition-based stats.
Identities = 63/120 (52%), Positives = 84/120 (70%), Gaps = 8/120 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
+SGGQKQRIAIARALI+ P IL+ DEATSALD +SERVVQEALD+ + TT+++AH
Sbjct 484 HMSGGQKQRIAIARALIKSPIILLLDEATSALDLESERVVQEALDN--ASVGRTTIVIAH 541
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLSTI+NAD+I VL H +V+ G+H++LM + G Y +V Q + + D+ S
Sbjct 542 RLSTIRNADIICVL-----HNGCIVETGSHDKLM-EIDGKYTSLVRLQQMKNEESCDNTS 595
Score = 108 bits (271), Expect = 8e-24, Method: Composition-based stats.
Identities = 59/109 (54%), Positives = 79/109 (72%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIAR +++ PSIL+ DEATSALD+QSERVVQ+AL+ ++ T++++AH
Sbjct 1121 QLSGGQKQRIAIARTILKNPSILLLDEATSALDSQSERVVQDALEHVM--VGKTSVVIAH 1178
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLM-RDTKGLYYHMVASQ 109
RLSTIQN D I VL+ VV+ GTH L+ + G Y+ +V+ Q
Sbjct 1179 RLSTIQNCDTIAVLDK-----GKVVESGTHASLLAKGPTGSYFSLVSLQ 1222
> cel:F22E10.1 pgp-12; P-GlycoProtein related family member (pgp-12);
K02021 putative ABC transport system ATP-binding protein
Length=1318
Score = 111 bits (278), Expect = 1e-24, Method: Composition-based stats.
Identities = 62/109 (56%), Positives = 77/109 (70%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
GQLSGGQKQRIAIARAL+R P IL+ DEATSALD++SER VQEALD T + +A
Sbjct 1214 GQLSGGQKQRIAIARALVRDPKILLLDEATSALDSESERAVQEALDR--AREGRTCITIA 1271
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRLS+IQN+DLIV ++ HG V + G H LM +G YY+++ Q
Sbjct 1272 HRLSSIQNSDLIVYID----HG-MVQEAGNHAHLM-SLRGKYYNLIKKQ 1314
Score = 111 bits (278), Expect = 1e-24, Method: Composition-based stats.
Identities = 72/175 (41%), Positives = 96/175 (54%), Gaps = 15/175 (8%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQR+AIAR LIR P +L+ DEATSALD QSE VVQ AL++ K TT+++AH
Sbjct 555 QLSGGQKQRVAIARTLIRDPKVLLLDEATSALDAQSESVVQSALNNAAKGR--TTIMIAH 612
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLSTI+ AD IV E +V+ G HE+L+ G Y +V +Q + D +D+
Sbjct 613 RLSTIREADKIVFFE-----NGVIVESGNHEELVA-LGGRYAKLVEAQKFKESDDIEDNG 666
Query 122 AVQETIPAPHEPEPADGAAVPTLQERRMSTDFVAASIAKASRSLAKTPNFRTYKL 176
HE E + L R++S S+A A + F T+ L
Sbjct 667 -------DEHEEETSTVGRHDRLSSRQVSFHKSCESLASADLEIGYASTFNTFTL 714
> ath:AT5G46540 PGP7; PGP7 (P-GLYCOPROTEIN 7); ATPase, coupled
to transmembrane movement of substances
Length=1248
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 62/136 (45%), Positives = 93/136 (68%), Gaps = 18/136 (13%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SER+VQ+AL L+ + TT++VAH
Sbjct 495 QLSGGQKQRIAIARAILKNPKILLLDEATSALDAESERIVQDALVKLMLSR--TTVVVAH 552
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RL+TI+ AD+I V++ V+++GTH+++++D +G Y SQ+V +G +
Sbjct 553 RLTTIRTADMIAVVQQ-----GKVIEKGTHDEMIKDPEGTY-----SQLVRLQEGSKKEE 602
Query 122 AVQETIPAPHEPEPAD 137
A+ + EPE +
Sbjct 603 AIDK------EPEKCE 612
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 76/106 (71%), Gaps = 7/106 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1144 QLSGGQKQRIAIARAILKDPKILLLDEATSALDAESERVVQDALDQVM--VNRTTVVVAH 1201
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVA 107
L+TI++AD+I V++ + + G HE LM + G Y +VA
Sbjct 1202 LLTTIKDADMIAVVK-----NGVIAESGRHETLMEISGGAYASLVA 1242
> dre:100136865 abcb4, zgc:172149; ATP-binding cassette, sub-family
B (MDR/TAP), member 4
Length=650
Score = 110 bits (276), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 60/109 (55%), Positives = 81/109 (74%), Gaps = 8/109 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQRIAIARAL+R P IL+ DEATSALD +SE +VQ ALD V+ + TT++VAH
Sbjct 535 QMSGGQKQRIAIARALVRNPKILLLDEATSALDAESETIVQAALDK-VRLGR-TTIVVAH 592
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQI 110
RLSTI+NAD+I + +V+ GTH++LM + KG+Y+ +V Q+
Sbjct 593 RLSTIRNADVI-----AGFQNGEIVELGTHDELM-ERKGIYHSLVNMQV 635
> cel:T21E8.2 pgp-7; P-GlycoProtein related family member (pgp-7);
K02021 putative ABC transport system ATP-binding protein
Length=1263
Score = 110 bits (275), Expect = 3e-24, Method: Composition-based stats.
Identities = 58/109 (53%), Positives = 79/109 (72%), Gaps = 8/109 (7%)
Query 1 GQLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVA 60
QLSGGQKQRIAIARA++R P +L+ DEATSALD+ SE+VVQ ALD+ + + +T++VA
Sbjct 1154 AQLSGGQKQRIAIARAILRNPKVLLLDEATSALDSDSEKVVQNALDT--ASERLSTVVVA 1211
Query 61 HRLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
HRL T+ NAD I VL+ V +QGTHE+L+R + +Y+ +V Q
Sbjct 1212 HRLLTVVNADSIAVLK-----NGKVAEQGTHEELLR-KRSIYWRLVQKQ 1254
Score = 105 bits (262), Expect = 8e-23, Method: Composition-based stats.
Identities = 57/118 (48%), Positives = 84/118 (71%), Gaps = 8/118 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIAR L+R P IL+ DEATSALD +SE+VVQ+AL++ + TT++VAH
Sbjct 519 QLSGGQKQRIAIARTLVRNPKILLLDEATSALDNESEQVVQKALEN--ASQGRTTIVVAH 576
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDD 119
RLSTI+NA I+V++ +V+ G H++L+ +G+Y +V +Q++ D ++
Sbjct 577 RLSTIRNASKIIVMQK-----GEIVEVGNHDELIA-KRGVYNDLVQAQLLESHDDHEE 628
> ath:AT4G18050 PGP9; PGP9 (P-GLYCOPROTEIN 9); ATPase, coupled
to transmembrane movement of substances
Length=1236
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 55/108 (50%), Positives = 83/108 (76%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQKQR+AIARA+++ P IL+ DEATSALD +SER+VQ+AL +L+ + TT++VAH
Sbjct 493 QMSGGQKQRLAIARAILKNPKILLLDEATSALDAESERIVQDALVNLM--SNRTTVVVAH 550
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RL+TI+ AD+I V+ H +V++GTH+++++D +G Y +V Q
Sbjct 551 RLTTIRTADVIAVV-----HQGKIVEKGTHDEMIQDPEGAYSQLVRLQ 593
Score = 110 bits (274), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 57/105 (54%), Positives = 78/105 (74%), Gaps = 7/105 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRIAIARA+++ P IL+ DEATSALD +SERVVQ+ALD ++ TT++VAH
Sbjct 1132 QLSGGQKQRIAIARAILKDPKILLLDEATSALDAESERVVQDALDRVM--VNRTTVVVAH 1189
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMV 106
RL+TI+NAD+I V++ + ++G HE LM+ + G Y +V
Sbjct 1190 RLTTIKNADVIAVVK-----NGVIAEKGRHETLMKISGGAYASLV 1229
> ath:AT1G10680 PGP10; PGP10 (P-GLYCOPROTEIN 10); ATPase, coupled
to transmembrane movement of substances
Length=1227
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 55/108 (50%), Positives = 81/108 (75%), Gaps = 7/108 (6%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
Q+SGGQ+QRIAIARA+++ P IL+ DEATSALD +SERVVQ+ALD L++ TT++VAH
Sbjct 1120 QMSGGQRQRIAIARAVLKNPEILLLDEATSALDVESERVVQQALDRLMR--DRTTVVVAH 1177
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQ 109
RLSTI+N+D+I V++ +++QG+H L+ + G Y +++ Q
Sbjct 1178 RLSTIKNSDMISVIQD-----GKIIEQGSHNILVENKNGPYSKLISLQ 1220
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 59/132 (44%), Positives = 88/132 (66%), Gaps = 7/132 (5%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLSGGQKQRI+I+RA+++ PSIL+ DEATSALD +SE++VQEALD ++ TT++VAH
Sbjct 499 QLSGGQKQRISISRAIVKNPSILLLDEATSALDAESEKIVQEALDRVM--VGRTTVVVAH 556
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVASQIVPKIDGDDDDS 121
RLST++NAD+I V+ G +++ G+H++L+ + G Y ++ Q + + S
Sbjct 557 RLSTVRNADIIAVV-----GGGKIIESGSHDELISNPDGAYSSLLRIQEAASPNLNHTPS 611
Query 122 AVQETIPAPHEP 133
T P P P
Sbjct 612 LPVSTKPLPELP 623
> mmu:27413 Abcb11, ABC16, Bsep, Lith1, PFIC2, PGY4, SPGP; ATP-binding
cassette, sub-family B (MDR/TAP), member 11; K05664
ATP-binding cassette, subfamily B (MDR/TAP), member 11
Length=1321
Score = 108 bits (271), Expect = 7e-24, Method: Composition-based stats.
Identities = 58/107 (54%), Positives = 77/107 (71%), Gaps = 8/107 (7%)
Query 2 QLSGGQKQRIAIARALIRWPSILIFDEATSALDTQSERVVQEALDSLVKTTKATTLIVAH 61
QLS G+KQRIAIARA++R P IL+ DEATSALDT+SE+ VQ ALD T +++AH
Sbjct 1218 QLSRGEKQRIAIARAIVRDPKILLLDEATSALDTESEKTVQLALDK--AREGRTCIVIAH 1275
Query 62 RLSTIQNADLIVVLEPSATHGATVVQQGTHEQLMRDTKGLYYHMVAS 108
RLSTIQN+D+I V+ V+++GTH++LM D KG YY +V +
Sbjct 1276 RLSTIQNSDIIAVMSQ-----GVVIEKGTHKKLM-DQKGAYYKLVIT 1316
Lambda K H
0.315 0.128 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40