bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_0939_orf2
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
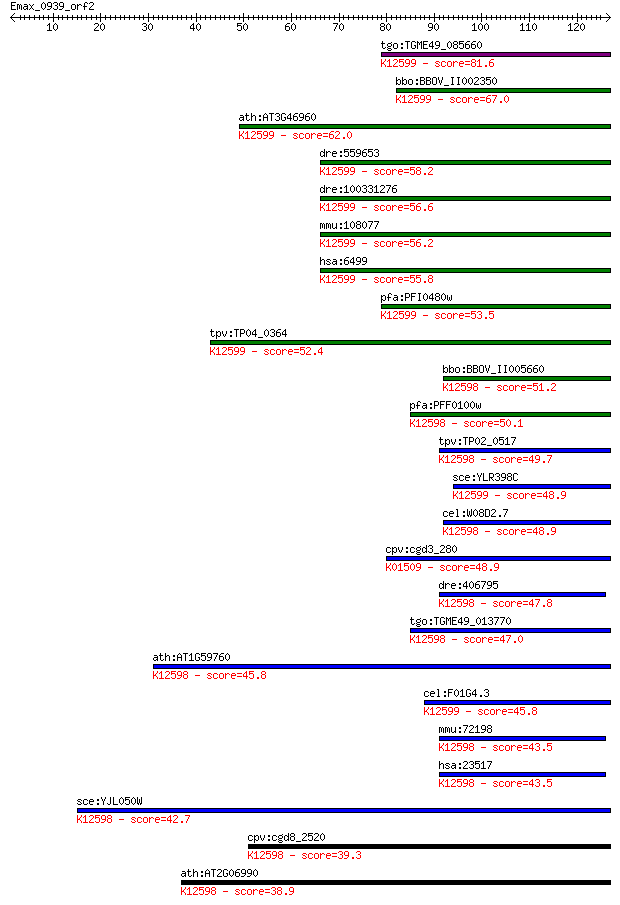
tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing pro... 81.6 5e-16
bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif p... 67.0 1e-11
ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/ ... 62.0 5e-10
dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidi... 58.2 7e-09
dre:100331276 superkiller viralicidic activity 2-like; K12599 ... 56.6 2e-08
mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;... 56.2 2e-08
hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superki... 55.8 4e-08
pfa:PFI0480w helicase with Zn-finger motif, putative; K12599 a... 53.5 2e-07
tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase ... 52.4 3e-07
bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing ... 51.2 8e-07
pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-)... 50.1 2e-06
tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA h... 49.7 2e-06
sce:YLR398C SKI2; Ski complex component and putative RNA helic... 48.9 4e-06
cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family m... 48.9 4e-06
cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase, DE... 48.9 4e-06
dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralic... 47.8 1e-05
tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependen... 47.0 2e-05
ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP... 45.8 3e-05
cel:F01G4.3 hypothetical protein; K12599 antiviral helicase SK... 45.8 3e-05
mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viral... 43.5 1e-04
hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118; s... 43.5 2e-04
sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, invo... 42.7 3e-04
cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP... 39.3 0.003
ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helic... 38.9 0.004
> tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing protein
; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1329
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 39/48 (81%), Positives = 41/48 (85%), Gaps = 0/48 (0%)
Query 79 ADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
DLSP + PLA+EF FPLDDFQKRAIL LEK Q VFVAAHTSAGKTVV
Sbjct 441 GDLSPLQVPLALEFPFPLDDFQKRAILHLEKYQTVFVAAHTSAGKTVV 488
> bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif
protein; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1113
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 31/45 (68%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 82 SPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+PE E L IE+ F LDDFQKRAI L K + VFVAAHTS+GKTVV
Sbjct 195 TPELEDLVIEYPFELDDFQKRAIYHLHKMKHVFVAAHTSSGKTVV 239
> ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1347
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/86 (45%), Positives = 50/86 (58%), Gaps = 11/86 (12%)
Query 49 AAAAAAAAAAAVTGAA--------AAAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQ 100
+A A + AVTG++ A D+ + +L P+ +AIEF F LD+FQ
Sbjct 309 SAKTAIMSEEAVTGSSDKQLRKEGWATKGDSQDIADRFYELVPD---MAIEFPFELDNFQ 365
Query 101 KRAILRLEKNQCVFVAAHTSAGKTVV 126
K AI LEK + VFVAAHTSAGKTVV
Sbjct 366 KEAICCLEKGESVFVAAHTSAGKTVV 391
> dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidic
activity 2 (S. cerevisiae homolog)-like; K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1230
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 34/61 (55%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A P D +SP AD A ++ F LD FQK+AILRLE + VFVAAHTSAGKTV
Sbjct 292 AIPVDISSP---CADFYKRIPDPAFKYPFELDVFQKQAILRLEAHDSVFVAAHTSAGKTV 348
Query 126 V 126
V
Sbjct 349 V 349
> dre:100331276 superkiller viralicidic activity 2-like; K12599
antiviral helicase SKI2 [EC:3.6.4.-]
Length=1235
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 38/61 (62%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A P D SP L P+ A ++ F D FQK+AIL LE++ VFVAAHTSAGKTV
Sbjct 284 AIPVDVTSPVGDFYRLIPQP---AFQWAFEPDVFQKQAILHLERHDSVFVAAHTSAGKTV 340
Query 126 V 126
V
Sbjct 341 V 341
> mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;
superkiller viralicidic activity 2-like (S. cerevisiae);
K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1244
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 38/61 (62%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A P D SP L P+ A ++ F D FQK+AIL LE++ VFVAAHTSAGKTV
Sbjct 281 AVPVDVTSPVGDFYRLIPQP---AFQWAFEPDVFQKQAILHLEQHDSVFVAAHTSAGKTV 337
Query 126 V 126
V
Sbjct 338 V 338
> hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superkiller
viralicidic activity 2-like (S. cerevisiae); K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1246
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 38/61 (62%), Gaps = 3/61 (4%)
Query 66 AAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
A P D SP L P+ A ++ F D FQK+AIL LE++ VFVAAHTSAGKTV
Sbjct 284 AIPVDATSPVGDFYRLIPQP---AFQWAFEPDVFQKQAILHLERHDSVFVAAHTSAGKTV 340
Query 126 V 126
V
Sbjct 341 V 341
> pfa:PFI0480w helicase with Zn-finger motif, putative; K12599
antiviral helicase SKI2 [EC:3.6.4.-]
Length=1373
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 34/49 (69%), Gaps = 1/49 (2%)
Query 79 ADLSP-EEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+D+S + L + + F LDDFQKR+I L + VFVAAHTSAGKT++
Sbjct 284 SDISNFNKNDLLLNYDFELDDFQKRSIKHLNNFKHVFVAAHTSAGKTLI 332
> tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1069
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 38/84 (45%), Gaps = 16/84 (19%)
Query 43 VAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKR 102
+ P PD A+ A PE L ++ F LDDFQK+
Sbjct 146 IVPKPDENKKYIRKKWAIVDDKEA----------------PELTDLIAQYPFELDDFQKK 189
Query 103 AILRLEKNQCVFVAAHTSAGKTVV 126
+I L + VFV+AHTSAGKTVV
Sbjct 190 SIHHLINGKHVFVSAHTSAGKTVV 213
> bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing
DEAD/DEAH box helicase family protein (EC:3.6.1.3); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=986
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 24/35 (68%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LDDFQKR+I LEK + V V AHTSAGKTVV
Sbjct 80 YPFTLDDFQKRSIECLEKGESVLVCAHTSAGKTVV 114
> pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1350
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/44 (56%), Positives = 33/44 (75%), Gaps = 2/44 (4%)
Query 85 EEPL--AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+EPL A + F LD FQK++I LE+N+ V V+AHTSAGKTV+
Sbjct 242 KEPLIPARTYKFELDTFQKKSIECLERNESVLVSAHTSAGKTVI 285
> tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA
helicase DOB1 [EC:3.6.4.13]
Length=1012
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 91 EFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
++ F LD+FQKR+I LE+++ V V AHTSAGKTVV
Sbjct 103 KYPFTLDEFQKRSIESLERDESVLVCAHTSAGKTVV 138
> sce:YLR398C SKI2; Ski complex component and putative RNA helicase,
mediates 3'-5' RNA degradation by the cytoplasmic exosome;
null mutants have superkiller phenotype of increased viral
dsRNAs and are synthetic lethal with mutations in 5'-3'
mRNA decay (EC:3.6.1.-); K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1287
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/33 (69%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 94 FPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
F LD FQK A+ LE+ VFVAAHTSAGKTVV
Sbjct 328 FELDTFQKEAVYHLEQGDSVFVAAHTSAGKTVV 360
> cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family
member (mtr-4); K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1026
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/35 (65%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQK+AIL ++ NQ V V+AHTSAGKTVV
Sbjct 122 YPFQLDAFQKQAILCIDNNQSVLVSAHTSAGKTVV 156
> cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase,
DEXDc+HELICc ; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1439
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 80 DLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
D S + E + +++ + LDDFQKRA++ + V VAAHTSAGKT V
Sbjct 112 DDSEDLENVVLKYPYELDDFQKRAVINIHNGDHVLVAAHTSAGKTAV 158
> dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralicidic
activity 2-like 2 (EC:3.6.4.13); K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1034
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/35 (62%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 91 EFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
E+ F LD FQ+ AIL ++ NQ V V+AHTSAGKTV
Sbjct 123 EYPFILDPFQREAILCIDNNQSVLVSAHTSAGKTV 157
> tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1206
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/42 (59%), Positives = 32/42 (76%), Gaps = 1/42 (2%)
Query 85 EEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+EP A ++ F LD FQ+R++L LE + V VAAHTSAGKTVV
Sbjct 225 KEP-ARQYKFRLDAFQQRSVLCLEAGESVLVAAHTSAGKTVV 265
> ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=988
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 44/98 (44%), Gaps = 3/98 (3%)
Query 31 GSAALESALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSP--EEEPL 88
GS +S + + PP + + D + P L+P +P
Sbjct 2 GSVKRKSVEESSDSAPPQKVQREDDSTQIINEELVGCVHDVSFPENYVP-LAPSVHNKPP 60
Query 89 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
A +F F LD FQ AI L+ + V V+AHTSAGKTVV
Sbjct 61 AKDFPFTLDSFQSEAIKCLDNGESVMVSAHTSAGKTVV 98
> cel:F01G4.3 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1266
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/39 (53%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 88 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+A ++ F LD FQ+ ++L +E+ + +FVAAHTSAGKTVV
Sbjct 285 MARKYPFSLDPFQQSSVLCMERGESLFVAAHTSAGKTVV 323
> mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viralicidic
activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1040
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 91 EFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
E+ F LD FQ+ AI ++ NQ V V+AHTSAGKTV
Sbjct 133 EYPFILDAFQREAIQCVDNNQSVLVSAHTSAGKTV 167
> hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118;
superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1042
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 91 EFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTV 125
E+ F LD FQ+ AI ++ NQ V V+AHTSAGKTV
Sbjct 135 EYPFILDAFQREAIQCVDNNQSVLVSAHTSAGKTV 169
> sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, involved
in nuclear RNA processing and degredation both as a component
of the TRAMP complex and in TRAMP independent processes;
member of the Dead-box family of helicases (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1073
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 42/112 (37%), Gaps = 26/112 (23%)
Query 15 AAAAAARQQQQQAAAGGSAALESALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASP 74
A+ + Q G L +R QVA PP+ A V A
Sbjct 95 ASKGLTNSETLQVEQDGKVRLSHQVRHQVALPPNYDYTPIAEHKRVNEART--------- 145
Query 75 STAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQ AI +++ + V V+AHTSAGKTVV
Sbjct 146 -----------------YPFTLDPFQDTAISCIDRGESVLVSAHTSAGKTVV 180
> cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1280
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query 51 AAAAAAAAAVTGAAAAAPTDTASPSTAAADLSPEEEPLAIEFGFPLDDFQKRAILRLEKN 110
+ + APTD S D + ++ P A E+ F LD FQ +I LE
Sbjct 58 SGTNCVHECIRPCGYVAPTD--SKLRVQYDENGKKIP-AKEYSFKLDTFQAVSIGCLEIG 114
Query 111 QCVFVAAHTSAGKTVV 126
+ V ++AHTSAGKT V
Sbjct 115 ESVLISAHTSAGKTCV 130
> ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helicase/
RNA helicase; K12598 ATP-dependent RNA helicase DOB1
[EC:3.6.4.13]
Length=995
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 43/95 (45%), Gaps = 7/95 (7%)
Query 37 SALRAQVAPPPDAAAAAAAAAAAVTGAAAAAPTDTASPSTAAADLSPEEEP-----LAIE 91
S LR+ P P+ + A A P D +P+ + P +A
Sbjct 20 SKLRSDETPTPEPRTKRRSLKRACV-HEVAVPND-YTPTKEETIHGTLDNPVFNGDMAKT 77
Query 92 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVV 126
+ F LD FQ ++ LE+ + + V+AHTSAGKT V
Sbjct 78 YPFKLDPFQSVSVACLERKESILVSAHTSAGKTAV 112
Lambda K H
0.314 0.121 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40