bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1088_orf3
Length=167
Score E
Sequences producing significant alignments: (Bits) Value
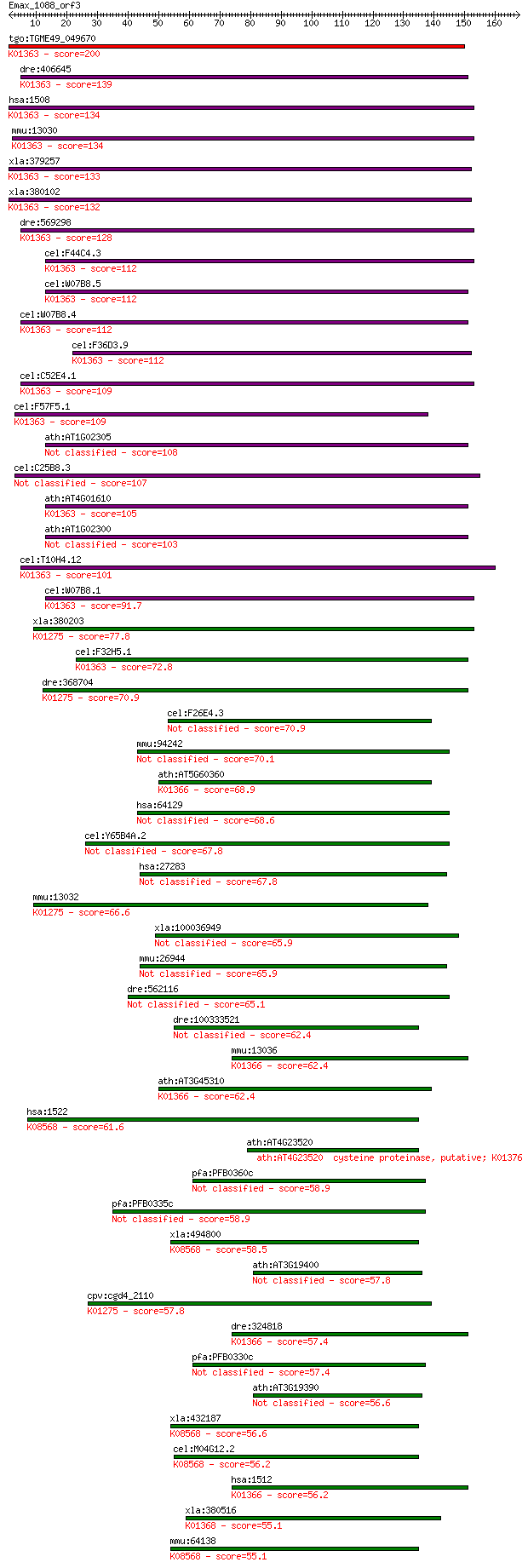
tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);... 200 2e-51
dre:406645 ctsba, MGC55862, ctsb, id:ibd1201, wu:fa13g05, wu:f... 139 5e-33
hsa:1508 CTSB, APPS, CPSB; cathepsin B (EC:3.4.22.1); K01363 c... 134 1e-31
mmu:13030 Ctsb, CB; cathepsin B (EC:3.4.22.1); K01363 cathepsi... 134 1e-31
xla:379257 ctsb, MGC53360, apps, cpsb; cathepsin B (EC:3.4.22.... 133 2e-31
xla:380102 cg10992; hypothetical protein MGC52983; K01363 cath... 132 4e-31
dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.... 128 1e-29
cel:F44C4.3 cpr-4; Cysteine PRotease related family member (cp... 112 4e-25
cel:W07B8.5 cpr-5; Cysteine PRotease related family member (cp... 112 6e-25
cel:W07B8.4 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 112 7e-25
cel:F36D3.9 cpr-2; Cysteine PRotease related family member (cp... 112 7e-25
cel:C52E4.1 cpr-1; Cysteine PRotease related family member (cp... 109 4e-24
cel:F57F5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 109 5e-24
ath:AT1G02305 cathepsin B-like cysteine protease, putative 108 7e-24
cel:C25B8.3 cpr-6; Cysteine PRotease related family member (cp... 107 2e-23
ath:AT4G01610 cathepsin B-like cysteine protease, putative; K0... 105 8e-23
ath:AT1G02300 cathepsin B-like cysteine protease, putative 103 3e-22
cel:T10H4.12 cpr-3; Cysteine PRotease related family member (c... 101 1e-21
cel:W07B8.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 91.7 1e-18
xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275 c... 77.8 2e-14
cel:F32H5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 72.8 6e-13
dre:368704 ctsc, cb912, ik:tdsubc_1h2, sb:cb146, wu:fb34g12, w... 70.9 2e-12
cel:F26E4.3 hypothetical protein 70.9 2e-12
mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,... 70.1 3e-12
ath:AT5G60360 AALP; AALP (Arabidopsis aleurain-like protease);... 68.9 8e-12
hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointersti... 68.6 8e-12
cel:Y65B4A.2 hypothetical protein 67.8 1e-11
hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen 67.8 2e-11
mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1... 66.6 3e-11
xla:100036949 ctsh; cathepsin H (EC:3.4.22.16) 65.9 5e-11
mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephriti... 65.9 6e-11
dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephr... 65.1 1e-10
dre:100333521 Cathepsin Z-like 62.4 6e-10
mmu:13036 Ctsh, AL022844; cathepsin H (EC:3.4.22.16); K01366 c... 62.4 7e-10
ath:AT3G45310 cysteine proteinase, putative; K01366 cathepsin ... 62.4 8e-10
hsa:1522 CTSZ, CTSX, FLJ17088; cathepsin Z (EC:3.4.18.1); K085... 61.6 1e-09
ath:AT4G23520 cysteine proteinase, putative; K01376 [EC:3.4.2... 59.7 4e-09
pfa:PFB0360c SERA-1; serine repeat antigen 1 (SERA-1) 58.9 7e-09
pfa:PFB0335c SERA-6, SERP; serine repeat antigen 6 (SERA-6) 58.9 8e-09
xla:494800 ctsz; cathepsin Z (EC:3.4.18.1); K08568 cathepsin X... 58.5 1e-08
ath:AT3G19400 cysteine proteinase, putative 57.8 2e-08
cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin C... 57.8 2e-08
dre:324818 ctsh, fc44c02, wu:fc44c02, zgc:85774; cathepsin H (... 57.4 2e-08
pfa:PFB0330c SERA-7; serine repeat antigen 7 (SERA-7) 57.4 2e-08
ath:AT3G19390 cysteine proteinase, putative / thiol protease, ... 56.6 4e-08
xla:432187 hypothetical protein MGC82409; K08568 cathepsin X [... 56.6 4e-08
cel:M04G12.2 cpz-2; CathePsin Z family member (cpz-2); K08568 ... 56.2 4e-08
hsa:1512 CTSH, ACC-4, ACC-5, CPSB, DKFZp686B24257, MGC1519, mi... 56.2 5e-08
xla:380516 ctss-a, MGC69026; cathepsin S (EC:3.4.22.27); K0136... 55.1 1e-07
mmu:64138 Ctsz, AI787083, AU019819, CTSX, D2Wsu143e; cathepsin... 55.1 1e-07
> tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);
K01363 cathepsin B [EC:3.4.22.1]
Length=572
Score = 200 bits (508), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 87/151 (57%), Positives = 111/151 (73%), Gaps = 2/151 (1%)
Query 1 EVPFCQHHSDGPYPQCDGPL--PKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGR 58
EVPFC HH+ P+P CD L K PKCRKDCEE Y VHPF D H A+++YS+ R
Sbjct 386 EVPFCAHHAKAPFPDCDATLVPRKTPKCRKDCEEQAYADNVHPFDQDTHKATSAYSLRSR 445
Query 59 DHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLA 118
D +K+++ +G ++GAF+V+EDFL YK GVY HV+G+P+GGHA+K+IG+G+E G +YW A
Sbjct 446 DDVKRDMMTHGPVSGAFMVYEDFLSYKSGVYKHVSGLPVGGHAIKIIGWGTENGEEYWHA 505
Query 119 VNSWNEYWGDKGTFKIQMGEAGIDKEFCGGE 149
VNSWN YWGD G FKI MG+ GID E GE
Sbjct 506 VNSWNTYWGDGGQFKIAMGQCGIDGEMVAGE 536
> dre:406645 ctsba, MGC55862, ctsb, id:ibd1201, wu:fa13g05, wu:fb34e12,
zgc:55862, zgc:65809, zgc:77181; cathepsin B, a; K01363
cathepsin B [EC:3.4.22.1]
Length=330
Score = 139 bits (350), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 73/149 (48%), Positives = 91/149 (61%), Gaps = 7/149 (4%)
Query 5 CQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIKK 63
C+HH +G P C G P C CE Y+ +K D HF TSYSV ++ I
Sbjct 185 CEHHVNGSRPPCSGEGGDTPNCDMKCEP-GYSPS---YKQDKHFGKTSYSVPSNQNSIMA 240
Query 64 ELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWN 123
EL +NG + GAF V+EDFL+YK GVY H++G P+GGHA+K++G+G E G YWLA NSWN
Sbjct 241 ELFKNGPVEGAFTVYEDFLLYKSGVYQHMSGSPVGGHAIKILGWGEENGVPYWLAANSWN 300
Query 124 EYWGDKGTFKIQMGE--AGIDKEFCGGEP 150
WGD G FKI GE GI+ E G P
Sbjct 301 TDWGDNGYFKILRGEDHCGIESEIVAGIP 329
> hsa:1508 CTSB, APPS, CPSB; cathepsin B (EC:3.4.22.1); K01363
cathepsin B [EC:3.4.22.1]
Length=339
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 72/155 (46%), Positives = 90/155 (58%), Gaps = 8/155 (5%)
Query 1 EVPFCQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRDH 60
+P C+HH +G P C G PKC K CE T +K D H+ SYSV +
Sbjct 183 SIPPCEHHVNGSRPPCTGE-GDTPKCSKICEPGYSPT----YKQDKHYGYNSYSVSNSEK 237
Query 61 -IKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAV 119
I E+ +NG + GAF V+ DFL+YK GVY HVTG MGGHA++++G+G E G YWL
Sbjct 238 DIMAEIYKNGPVEGAFSVYSDFLLYKSGVYQHVTGEMMGGHAIRILGWGVENGTPYWLVA 297
Query 120 NSWNEYWGDKGTFKIQMGE--AGIDKEFCGGEPRV 152
NSWN WGD G FKI G+ GI+ E G PR
Sbjct 298 NSWNTDWGDNGFFKILRGQDHCGIESEVVAGIPRT 332
> mmu:13030 Ctsb, CB; cathepsin B (EC:3.4.22.1); K01363 cathepsin
B [EC:3.4.22.1]
Length=339
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 74/154 (48%), Positives = 90/154 (58%), Gaps = 8/154 (5%)
Query 2 VPFCQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGR-DH 60
+P C+HH +G P C G P+C K CE Y+ +K+D HF TSYSV
Sbjct 184 IPPCEHHVNGSRPPCTGE-GDTPRCNKSCE-AGYSPS---YKEDKHFGYTSYSVSNSVKE 238
Query 61 IKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVN 120
I E+ +NG + GAF VF DFL YK GVY H G MGGHA++++G+G E G YWLA N
Sbjct 239 IMAEIYKNGPVEGAFTVFSDFLTYKSGVYKHEAGDMMGGHAIRILGWGVENGVPYWLAAN 298
Query 121 SWNEYWGDKGTFKIQMGE--AGIDKEFCGGEPRV 152
SWN WGD G FKI GE GI+ E G PR
Sbjct 299 SWNLDWGDNGFFKILRGENHCGIESEIVAGIPRT 332
> xla:379257 ctsb, MGC53360, apps, cpsb; cathepsin B (EC:3.4.22.1);
K01363 cathepsin B [EC:3.4.22.1]
Length=333
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 71/154 (46%), Positives = 90/154 (58%), Gaps = 7/154 (4%)
Query 1 EVPFCQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRD 59
+P C+HH +G P C G PKC K CEE Y+ + D HF +TSY V
Sbjct 183 SIPPCEHHVNGSRPACKGEEGDTPKCVKQCEE-GYSPA---YGTDKHFGTTSYGVPTSEK 238
Query 60 HIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAV 119
I E+ +NG + GAFLV+ DF +YK GVY H TG +GGHA+K++G+G E G YWL
Sbjct 239 EIMAEIYKNGPVEGAFLVYADFPLYKSGVYQHETGEELGGHAIKILGWGVENGTPYWLCA 298
Query 120 NSWNEYWGDKGTFKIQMGE--AGIDKEFCGGEPR 151
NSWN WGD G FKI G+ GI+ E G P+
Sbjct 299 NSWNTDWGDNGFFKILRGKDHCGIESEIVAGVPK 332
> xla:380102 cg10992; hypothetical protein MGC52983; K01363 cathepsin
B [EC:3.4.22.1]
Length=333
Score = 132 bits (333), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 70/154 (45%), Positives = 91/154 (59%), Gaps = 7/154 (4%)
Query 1 EVPFCQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRD- 59
+P C+HH +G P C G PKC K CEE YT + D HF +TSY V +
Sbjct 183 SIPPCEHHVNGSRPSCKGEEGDTPKCMKTCEE-GYTPA---YGSDKHFGATSYGVPSSEK 238
Query 60 HIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAV 119
I ++ +NG + GAF+V+ DF +YK GVY H TG +GGHA+K++G+G E G YWL
Sbjct 239 EIMADIYKNGPVEGAFVVYADFPLYKSGVYQHETGEELGGHAIKILGWGVENGTPYWLCA 298
Query 120 NSWNEYWGDKGTFKIQMGE--AGIDKEFCGGEPR 151
NSWN WGD G FKI G+ GI+ E G P+
Sbjct 299 NSWNTDWGDNGFFKILRGKDHCGIESEVVAGIPK 332
> dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.22.1]
Length=326
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 69/151 (45%), Positives = 88/151 (58%), Gaps = 8/151 (5%)
Query 5 CQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIKK 63
C+HH +G P C G PKC C +Y+ P+K D HF S Y+V + I
Sbjct 181 CEHHVNGTRPPCSGE-QDTPKCTGVCIP-KYSV---PYKQDKHFGSKVYNVPSDQQQIMT 235
Query 64 ELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWN 123
EL NG + AF V+EDF +YK GVY H+TG +GGHAVK++G+G E G +WL NSWN
Sbjct 236 ELYTNGPVEAAFTVYEDFPLYKSGVYQHLTGSALGGHAVKILGWGEENGTPFWLVANSWN 295
Query 124 EYWGDKGTFKIQMG--EAGIDKEFCGGEPRV 152
WGD G FKI G E GI+ E G P++
Sbjct 296 SDWGDNGYFKILRGHDECGIESEMVAGLPKL 326
> cel:F44C4.3 cpr-4; Cysteine PRotease related family member (cpr-4);
K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 112 bits (281), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 57/143 (39%), Positives = 79/143 (55%), Gaps = 6/143 (4%)
Query 13 YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGR-DHIKKELKENGTL 71
+P C P C C Y + D HF ST+Y+V + I+ E+ +G +
Sbjct 196 WPSCPDDGYDTPACVNKCTNKNYNVA---YTADKHFGSTAYAVGKKVSQIQAEIIAHGPV 252
Query 72 TGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGT 131
AF V+EDF YK GVY H TG +GGHA++++G+G++ G YWL NSWN WG+ G
Sbjct 253 EAAFTVYEDFYQYKTGVYVHTTGQELGGHAIRILGWGTDNGTPYWLVANSWNVNWGENGY 312
Query 132 FKIQMG--EAGIDKEFCGGEPRV 152
F+I G E GI+ GG P+V
Sbjct 313 FRIIRGTNECGIEHAVVGGVPKV 335
> cel:W07B8.5 cpr-5; Cysteine PRotease related family member (cpr-5);
K01363 cathepsin B [EC:3.4.22.1]
Length=344
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 60/142 (42%), Positives = 79/142 (55%), Gaps = 7/142 (4%)
Query 13 YPQCDGPLPKAPKCRKDC-EEVEYTTKVHPFKDDLHFASTSYSVEGR-DHIKKELKENGT 70
+P C PKC C + Y T P+ D HF ST+Y+V + + I+ E+ NG
Sbjct 200 WPACPEDTEPTPKCVDSCTSKNNYAT---PYLQDKHFGSTAYAVGKKVEQIQTEILTNGP 256
Query 71 LTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKG 130
+ AF V+EDF Y GVY H G +GGHAVK++G+G + G YWL NSWN WG+KG
Sbjct 257 IEVAFTVYEDFYQYTTGVYVHTAGASLGGHAVKILGWGVDNGTPYWLVANSWNVAWGEKG 316
Query 131 TFKIQMG--EAGIDKEFCGGEP 150
F+I G E GI+ G P
Sbjct 317 YFRIIRGLNECGIEHSAVAGIP 338
> cel:W07B8.4 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 112 bits (279), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 58/150 (38%), Positives = 84/150 (56%), Gaps = 6/150 (4%)
Query 5 CQHHSDG-PYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIK 62
C DG +P+C + PKC C + P+ D HF +++Y++ I+
Sbjct 182 CGETIDGVTWPECPMKISDTPKCEHHC--TGNNSYPIPYDQDKHFGASAYAIGRSAKQIQ 239
Query 63 KELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSW 122
E+ +G + F+V+EDF +YK G+Y HV G +GGHAVK++G+G + G YWLA NSW
Sbjct 240 TEILAHGPVEVGFIVYEDFYLYKTGIYTHVAGGELGGHAVKMLGWGVDNGTPYWLAANSW 299
Query 123 NEYWGDKGTFKIQMG--EAGIDKEFCGGEP 150
N WG+KG F+I G E GI+ G P
Sbjct 300 NTVWGEKGYFRILRGVDECGIESAAVAGMP 329
> cel:F36D3.9 cpr-2; Cysteine PRotease related family member (cpr-2);
K01363 cathepsin B [EC:3.4.22.1]
Length=344
Score = 112 bits (279), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 56/133 (42%), Positives = 79/133 (59%), Gaps = 7/133 (5%)
Query 22 KAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIKKELKENGTLTGAFLVFED 80
+ P CR C+ TT + +D ++ +++Y V I+ ++ NG + AF+V+ED
Sbjct 216 QTPPCRLSCQPGYRTT----YTNDKNYGNSAYPVPRTVAAIQADIYYNGPVVAAFIVYED 271
Query 81 FLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGTFKIQMG--E 138
F YK G+Y H+ G GGHAVK+IG+G+E G YWLAVNSW WG+ GTF+I G E
Sbjct 272 FEKYKSGIYRHIAGRSKGGHAVKLIGWGTERGTPYWLAVNSWGSQWGESGTFRILRGVDE 331
Query 139 AGIDKEFCGGEPR 151
GI+ G PR
Sbjct 332 CGIESRIVAGLPR 344
> cel:C52E4.1 cpr-1; Cysteine PRotease related family member (cpr-1);
K01363 cathepsin B [EC:3.4.22.1]
Length=329
Score = 109 bits (272), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 58/151 (38%), Positives = 82/151 (54%), Gaps = 7/151 (4%)
Query 5 CQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIKK 63
C+ + P + P K P C C+ Y+T + D HF ++Y+V + I+
Sbjct 183 CKPYPIAPCTSGNCPESKTPSCSMSCQS-GYST---AYAKDKHFGVSAYAVPKNAASIQA 238
Query 64 ELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWN 123
E+ NG + AF V+EDF YK GVY H G +GGHA+K+IG+G+E G YWL NSW
Sbjct 239 EIYANGPVEAAFSVYEDFYKYKSGVYKHTAGKYLGGHAIKIIGWGTESGSPYWLVANSWG 298
Query 124 EYWGDKGTFKIQMG--EAGIDKEFCGGEPRV 152
WG+ G FKI G + GI+ G+ +V
Sbjct 299 VNWGESGFFKIYRGDDQCGIESAVVAGKAKV 329
> cel:F57F5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=400
Score = 109 bits (272), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 56/137 (40%), Positives = 79/137 (57%), Gaps = 6/137 (4%)
Query 3 PFCQHHSDGP-YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRD-H 60
P C+HH +G Y C + KC + C+ T ++ DLHF ++Y+V +
Sbjct 251 PPCEHHVNGTHYKPCPSNMYPTDKCERSCQAGYALT----YQQDLHFGQSAYAVSKKAAE 306
Query 61 IKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVN 120
I+KE+ +G + AF V+EDF Y GVY H G +GGHAVK++G+G + G YWL N
Sbjct 307 IQKEIMTHGPVEVAFTVYEDFEHYSGGVYVHTAGASLGGHAVKMLGWGVDNGTPYWLCAN 366
Query 121 SWNEYWGDKGTFKIQMG 137
SWNE WG+ G F+I G
Sbjct 367 SWNEDWGENGYFRIIRG 383
> ath:AT1G02305 cathepsin B-like cysteine protease, putative
Length=362
Score = 108 bits (271), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 60/142 (42%), Positives = 82/142 (57%), Gaps = 10/142 (7%)
Query 13 YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGR-DHIKKELKENGTL 71
+P C+ P PKC + C + +++ H+ ++Y V D I E+ +NG +
Sbjct 207 HPGCEPAYP-TPKCARKC-----VSGNQLWRESKHYGVSAYKVRSHPDDIMAEVYKNGPV 260
Query 72 TGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEGRDYWLAVNSWNEYWGDKG 130
AF V+EDF YK GVY H+TG +GGHAVK+IG+G S++G DYWL N WN WGD G
Sbjct 261 EVAFTVYEDFAHYKSGVYKHITGTNIGGHAVKLIGWGTSDDGEDYWLLANQWNRSWGDDG 320
Query 131 TFKIQMG--EAGIDKEFCGGEP 150
FKI+ G E GI+ G P
Sbjct 321 YFKIRRGTNECGIEHGVVAGLP 342
> cel:C25B8.3 cpr-6; Cysteine PRotease related family member (cpr-6)
Length=379
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 66/156 (42%), Positives = 89/156 (57%), Gaps = 7/156 (4%)
Query 3 PFCQHHSDGP-YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDH 60
P C+HHS + C L PKC K C +YT K + +D F +++Y V + +
Sbjct 209 PPCEHHSKKTHFDPCPHDLYPTPKCEKKCVS-DYTDKT--YSEDKFFGASAYGVKDDVEA 265
Query 61 IKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVN 120
I+KEL +G L AF V+EDFL Y GVY H G GGHAVK+IG+G ++G YW N
Sbjct 266 IQKELMTHGPLEIAFEVYEDFLNYDGGVYVHTGGKLGGGHAVKLIGWGIDDGIPYWTVAN 325
Query 121 SWNEYWGDKGTFKIQMG--EAGIDKEFCGGEPRVPT 154
SWN WG+ G F+I G E GI+ GG P++ +
Sbjct 326 SWNTDWGEDGFFRILRGVDECGIESGVVGGIPKLNS 361
> ath:AT4G01610 cathepsin B-like cysteine protease, putative;
K01363 cathepsin B [EC:3.4.22.1]
Length=359
Score = 105 bits (262), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 59/142 (41%), Positives = 82/142 (57%), Gaps = 10/142 (7%)
Query 13 YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRDH-IKKELKENGTL 71
+P C+ P PKC + C + + + H++ ++Y+V+ I E+ +NG +
Sbjct 204 HPGCEPAYP-TPKCSRKC-----VSDNKLWSESKHYSVSTYTVKSNPQDIMAEVYKNGPV 257
Query 72 TGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEGRDYWLAVNSWNEYWGDKG 130
+F V+EDF YK GVY H+TG +GGHAVK+IG+G S EG DYWL N WN WGD G
Sbjct 258 EVSFTVYEDFAHYKSGVYKHITGSNIGGHAVKLIGWGTSSEGEDYWLMANQWNRGWGDDG 317
Query 131 TFKIQMG--EAGIDKEFCGGEP 150
F I+ G E GI+ E G P
Sbjct 318 YFMIRRGTNECGIEDEPVAGLP 339
> ath:AT1G02300 cathepsin B-like cysteine protease, putative
Length=379
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 57/142 (40%), Positives = 81/142 (57%), Gaps = 10/142 (7%)
Query 13 YPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVE-GRDHIKKELKENGTL 71
+P C+ P PKC + C ++ + + H+ +Y + I E+ +NG +
Sbjct 224 HPGCEPTYP-TPKCERKC-----VSRNQLWGESKHYGVGAYRINPDPQDIMAEVYKNGPV 277
Query 72 TGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEGRDYWLAVNSWNEYWGDKG 130
AF V+EDF YK GVY ++TG +GGHAVK+IG+G S++G DYWL N WN WGD G
Sbjct 278 EVAFTVYEDFAHYKSGVYKYITGTKIGGHAVKLIGWGTSDDGEDYWLLANQWNRSWGDDG 337
Query 131 TFKIQMG--EAGIDKEFCGGEP 150
FKI+ G E GI++ G P
Sbjct 338 YFKIRRGTNECGIEQSVVAGLP 359
> cel:T10H4.12 cpr-3; Cysteine PRotease related family member
(cpr-3); K01363 cathepsin B [EC:3.4.22.1]
Length=370
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 56/160 (35%), Positives = 87/160 (54%), Gaps = 9/160 (5%)
Query 5 CQHHSDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV---EGRDHI 61
C +S P + + P P C+ C+ + K +K D H+ +++Y V + I
Sbjct 190 CMPYSFAPCTK-NCPESTTPSCKTTCQS---SYKTEEYKKDKHYGASAYKVTTTKSVTEI 245
Query 62 KKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNS 121
+ E+ G + ++ V+EDF YK GVYH+ +G +GGHAVK+IG+G E G DYWL NS
Sbjct 246 QTEIYHYGPVEASYKVYEDFYHYKSGVYHYTSGKLVGGHAVKIIGWGVENGVDYWLIANS 305
Query 122 WNEYWGDKGTFKIQMG--EAGIDKEFCGGEPRVPTNLNPF 159
W +G+KG FKI+ G E I+ G ++ T+ +
Sbjct 306 WGTSFGEKGFFKIRRGTNECQIEGNVVAGIAKLGTHSETY 345
> cel:W07B8.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 47/143 (32%), Positives = 71/143 (49%), Gaps = 5/143 (3%)
Query 13 YPQCDGPLPKAPKCRKDC-EEVEYTTKVHPFKDDLHFASTSYSVEGRDHIKKELKENGTL 71
YP C P C K C + Y + KD + S + I+ ++ NG +
Sbjct 194 YPACTNTTSPTPSCEKKCTSRIGYPIDID--KDRHYGVSVDQLPNSQIEIQSDVMLNGPI 251
Query 72 TGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGT 131
F V++DFL Y G+Y H+TG G +V++IG+G +G YWL NSW WG+ GT
Sbjct 252 QATFEVYDDFLQYTTGIYVHLTGNKQGHLSVRIIGWGVWQGVPYWLCANSWGRQWGENGT 311
Query 132 FKIQMG--EAGIDKEFCGGEPRV 152
F++ G E G++ G P++
Sbjct 312 FRVLRGTNECGLESNCVSGMPKL 334
> xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275
cathepsin C [EC:3.4.14.1]
Length=458
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 53/155 (34%), Positives = 76/155 (49%), Gaps = 24/155 (15%)
Query 9 SDGPYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRDHIKKELKEN 68
SD PY D P KD + YT + H+ Y ++K EL
Sbjct 315 SDFPYIGSDSPC-----TLKDSYQRYYTA-------EYHYVGGFYGGCNEAYMKLELVLG 362
Query 69 GTLTGAFLVFEDFLMYKKGVYHHVTGIP-------MGGHAVKVIGYGSEE--GRDYWLAV 119
G L+ AF V++DF+ Y+ GVYHH TG+ + HAV ++GYG+++ G YW+
Sbjct 363 GPLSVAFEVYDDFIHYRSGVYHH-TGLQDKFNPFQLTNHAVLLVGYGTDQQTGEKYWIVK 421
Query 120 NSWNEYWGDKGTFKIQMG--EAGIDKEFCGGEPRV 152
NSW E WG+KG F+I+ G E I+ P +
Sbjct 422 NSWGESWGEKGFFRIRRGSDECAIESIAVSANPII 456
> cel:F32H5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=356
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 63/131 (48%), Gaps = 5/131 (3%)
Query 23 APKCRKDCEEVEYTTKVHPFKDDLHFASTSYSV-EGRDHIKKELKENGTLTGAFLVFEDF 81
P C + C T +K D HF Y+V + I+ E+ NG + +F++++DF
Sbjct 222 TPTCEEHC--TSNITWPIAYKQDKHFGKAHYNVGKKMTDIQIEIMTNGPVIASFIIYDDF 279
Query 82 LMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGTFKIQMG--EA 139
YK G+Y H G GG K+IG+G + G YWL V+ W +G+ G + G E
Sbjct 280 WDYKTGIYVHTAGDQEGGMDTKIIGWGVDNGVPYWLCVHQWGTDFGENGFVRFLRGVNEV 339
Query 140 GIDKEFCGGEP 150
I+ + P
Sbjct 340 NIEHQVLAALP 350
> dre:368704 ctsc, cb912, ik:tdsubc_1h2, sb:cb146, wu:fb34g12,
wu:fj58d01; cathepsin C (EC:3.4.14.1); K01275 cathepsin C [EC:3.4.14.1]
Length=455
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/150 (31%), Positives = 66/150 (44%), Gaps = 24/150 (16%)
Query 12 PYPQCDGPLPKAPKCRKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRDHIKKELKENGTL 71
PY D P KC K + D H+ Y + EL +NG +
Sbjct 315 PYTGSDSPCNLPAKCTKY------------YASDYHYVGGFYGGCSESAMMLELVKNGPM 362
Query 72 TGAFLVFEDFLMYKKGVYHHVTGI-------PMGGHAVKVIGYGS--EEGRDYWLAVNSW 122
A V+ DF+ YK+G+YHH TG+ + HAV ++GYG + G YW+ NSW
Sbjct 363 GVALEVYPDFMNYKEGIYHH-TGLRDANNPFELTNHAVLLVGYGQCHKTGEKYWIVKNSW 421
Query 123 NEYWGDKGTFKIQMG--EAGIDKEFCGGEP 150
WG+ G F+I+ G E I+ P
Sbjct 422 GSGWGENGFFRIRRGTDECAIESIAVAATP 451
> cel:F26E4.3 hypothetical protein
Length=452
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 51/99 (51%), Gaps = 13/99 (13%)
Query 53 YSVEGRDH-IKKELKENGTLTGAFLVFEDFLMYKKGVYHH--------VTGIPMGGHAVK 103
Y V R+ I+ EL NG + F+V EDF MY GVY H + + G H+V+
Sbjct 318 YKVSSREEDIQTELMTNGPVQATFVVHEDFFMYAGGVYQHSDLAAQKGASSVAEGYHSVR 377
Query 104 VIGYGSEEGR----DYWLAVNSWNEYWGDKGTFKIQMGE 138
V+G+G + YWL NSW WG+ G FK+ GE
Sbjct 378 VLGWGVDHSTGKPIKYWLCANSWGTQWGEDGYFKVLRGE 416
> mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,
Tinagl; tubulointerstitial nephritis antigen-like 1
Length=466
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 63/117 (53%), Gaps = 16/117 (13%)
Query 43 KDDLHFASTSYSVEGRDH--IKKELKENGTLTGAFLVFEDFLMYKKGVYHHV---TGIP- 96
+D++ + +Y + G D I KEL ENG + V EDF +Y++G+Y H G P
Sbjct 333 SNDIYQVTPAYRL-GSDEKEIMKELMENGPVQALMEVHEDFFLYQRGIYSHTPVSQGRPE 391
Query 97 ----MGGHAVKVIGYGSE---EGRD--YWLAVNSWNEYWGDKGTFKIQMGEAGIDKE 144
G H+VK+ G+G E +GR YW A NSW +WG++G F+I G D E
Sbjct 392 QYRRHGTHSVKITGWGEETLPDGRTIKYWTAANSWGPWWGERGHFRIVRGTNECDIE 448
> ath:AT5G60360 AALP; AALP (Arabidopsis aleurain-like protease);
cysteine-type peptidase; K01366 cathepsin H [EC:3.4.22.16]
Length=357
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query 50 STSYSVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVY--HHVTGIPMG-GHAVKVIG 106
S + ++ D +K + ++ AF V F +YK GVY H PM HAV +G
Sbjct 252 SVNITLGAEDELKHAVGLVRPVSIAFEVIHSFRLYKSGVYTDSHCGSTPMDVNHAVLAVG 311
Query 107 YGSEEGRDYWLAVNSWNEYWGDKGTFKIQMGE 138
YG E+G YWL NSW WGDKG FK++MG+
Sbjct 312 YGVEDGVPYWLIKNSWGADWGDKGYFKMEMGK 343
> hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointerstitial
nephritis antigen-like 1
Length=436
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 59/116 (50%), Gaps = 14/116 (12%)
Query 43 KDDLHFASTSYSVEGRD-HIKKELKENGTLTGAFLVFEDFLMYKKGVYHHV---TGIP-- 96
+D++ + Y + D I KEL ENG + V EDF +YK G+Y H G P
Sbjct 303 NNDIYQVTPVYRLGSNDKEIMKELMENGPVQALMEVHEDFFLYKGGIYSHTPVSLGRPER 362
Query 97 ---MGGHAVKVIGYGSE---EGRD--YWLAVNSWNEYWGDKGTFKIQMGEAGIDKE 144
G H+VK+ G+G E +GR YW A NSW WG++G F+I G D E
Sbjct 363 YRRHGTHSVKITGWGEETLPDGRTLKYWTAANSWGPAWGERGHFRIVRGVNECDIE 418
> cel:Y65B4A.2 hypothetical protein
Length=421
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 55/166 (33%), Positives = 76/166 (45%), Gaps = 53/166 (31%)
Query 26 CRKDCEEVEYTTKVHPFKDDLHFASTSYSV------------------------------ 55
C K C+ + Y K +++D HFA+ +YS+
Sbjct 258 CMKRCQNIYYQQK---YEEDKHFATFAYSMYPRSMTVSPDGKERVKVPTIIGHFNDKKTE 314
Query 56 -----EGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGG--------HAV 102
E RD IKKE+ G T AF V E+FL Y GV+ P G H V
Sbjct 315 KLNVTEYRDIIKKEILLYGPTTMAFPVPEEFLHYSSGVFR---PYPTDGFDDRIVYWHVV 371
Query 103 KVIGYG-SEEGRDYWLAVNSWNEYWGDKGTFKIQ---MGEAGIDKE 144
++IG+G S++G YWLAVNS+ +WGD G FKI M + G++ E
Sbjct 372 RLIGWGESDDGTHYWLAVNSFGNHWGDNGLFKINTDDMEKYGLEYE 417
> hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen
Length=476
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 59/116 (50%), Gaps = 16/116 (13%)
Query 44 DDLHFASTSYSVEGRD-HIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGI------- 95
+ ++ S Y V + I KE+ +NG + V EDF YK G+Y HVT
Sbjct 346 NRIYQCSPPYRVSSNETEIMKEIMQNGPVQAIMQVREDFFHYKTGIYRHVTSTNKESEKY 405
Query 96 -PMGGHAVKVIGYGSEEG-----RDYWLAVNSWNEYWGDKGTFKIQMG--EAGIDK 143
+ HAVK+ G+G+ G +W+A NSW + WG+ G F+I G E+ I+K
Sbjct 406 RKLQTHAVKLTGWGTLRGAQGQKEKFWIAANSWGKSWGENGYFRILRGVNESDIEK 461
> mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=462
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 54/156 (34%), Positives = 74/156 (47%), Gaps = 28/156 (17%)
Query 9 SDGPYPQ-CDGPLPK--APKCRKDCEEVE-----YTTKVHP----------FKDDLHFAS 50
S PY Q CDG P A K +D VE YT K P + D ++
Sbjct 289 SCSPYAQGCDGGFPYLIAGKYAQDFGVVEESCFPYTAKDSPCKPRENCLRYYSSDYYYVG 348
Query 51 TSYSVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIP-------MGGHAVK 103
Y +K EL ++G + AF V +DFL Y G+YHH TG+ + HAV
Sbjct 349 GFYGGCNEALMKLELVKHGPMAVAFEVHDDFLHYHSGIYHH-TGLSDPFNPFELTNHAVL 407
Query 104 VIGYGSE--EGRDYWLAVNSWNEYWGDKGTFKIQMG 137
++GYG + G +YW+ NSW WG+ G F+I+ G
Sbjct 408 LVGYGRDPVTGIEYWIIKNSWGSNWGESGYFRIRRG 443
> xla:100036949 ctsh; cathepsin H (EC:3.4.22.16)
Length=319
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 58/106 (54%), Gaps = 13/106 (12%)
Query 49 ASTSYSVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG 108
S Y + +++ + ++G +T F V EDF+ Y KG++ P HA+ V+GYG
Sbjct 208 VSKYYILPDEENMASSVAKDGPITVGFAVAEDFMFYSKGIFDGECA-PSPNHAIIVVGYG 266
Query 109 S-------EEGRDYWLAVNSWNEYWGDKGTFKIQMGEAGIDKEFCG 147
+ ++G DYW+ NSW E+WG++G KIQ +K+ CG
Sbjct 267 TLHCEDGEDDGEDYWIIKNSWGEHWGEEGFGKIQR-----NKDMCG 307
> mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephritis
antigen
Length=475
Score = 65.9 bits (159), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 58/116 (50%), Gaps = 16/116 (13%)
Query 44 DDLHFASTSYSVEGRD-HIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGI------- 95
+ ++ S Y V + I +E+ +NG + V EDF YK G+Y HV
Sbjct 345 NRIYQCSPPYRVSSNETEIMREIIQNGPVQAIMQVHEDFFYYKTGIYRHVVSTNEEPEKY 404
Query 96 -PMGGHAVKVIGYGSEEG-----RDYWLAVNSWNEYWGDKGTFKIQMG--EAGIDK 143
+ HAVK+ G+G+ G +W+A NSW + WG+ G F+I G E+ I+K
Sbjct 405 KKLRTHAVKLTGWGTLRGARGKKEKFWIAANSWGKSWGENGYFRILRGVNESDIEK 460
> dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephritis
antigen-like 1
Length=471
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 57/119 (47%), Gaps = 14/119 (11%)
Query 40 HPFKDDLHFASTSYSVE-GRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTG---- 94
H + +D++ ++ Y + + I KE+ +NG + V EDF +YK G++ H
Sbjct 327 HSYHNDIYQSTPPYRLSTNENEIMKEIMDNGPVQAIMEVHEDFFVYKSGIFRHTDVNYHK 386
Query 95 ----IPMGGHAVKVIGYGSEE-----GRDYWLAVNSWNEYWGDKGTFKIQMGEAGIDKE 144
H+V++ G+G E R YW+ NSW + WG+ G F+I G D E
Sbjct 387 PSQYRKHATHSVRITGWGEERDYSGRTRKYWIGANSWGKNWGEDGYFRIARGVNECDIE 445
> dre:100333521 Cathepsin Z-like
Length=267
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 47/81 (58%), Gaps = 1/81 (1%)
Query 55 VEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEGR 113
+ GRD +K E+ +NG ++ A + + Y GV+ + M H + V G+G +E+G
Sbjct 158 ISGRDRMKAEIFKNGPISCAIMATKGLEAYDGGVFAEFHILSMPNHIISVAGWGVTEDGT 217
Query 114 DYWLAVNSWNEYWGDKGTFKI 134
+YW+ NSW E+WG+ G +I
Sbjct 218 EYWIVRNSWGEFWGESGWARI 238
> mmu:13036 Ctsh, AL022844; cathepsin H (EC:3.4.22.16); K01366
cathepsin H [EC:3.4.22.16]
Length=333
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/81 (43%), Positives = 42/81 (51%), Gaps = 5/81 (6%)
Query 74 AFLVFEDFLMYKKGVYH----HVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDK 129
AF V EDFLMYK GVY H T + HAV +GYG + G YW+ NSW WG+
Sbjct 250 AFEVTEDFLMYKSGVYSSKSCHKTPDKVN-HAVLAVGYGEQNGLLYWIVKNSWGSQWGEN 308
Query 130 GTFKIQMGEAGIDKEFCGGEP 150
G F I+ G+ C P
Sbjct 309 GYFLIERGKNMCGLAACASYP 329
> ath:AT3G45310 cysteine proteinase, putative; K01366 cathepsin
H [EC:3.4.22.16]
Length=357
Score = 62.4 bits (150), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 50/92 (54%), Gaps = 3/92 (3%)
Query 50 STSYSVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVT--GIPMG-GHAVKVIG 106
S + ++ D +K + ++ AF V +F YKKGV+ T PM HAV +G
Sbjct 252 SVNITLGAEDELKHAVGLVRPVSVAFEVVHEFRFYKKGVFTSNTCGNTPMDVNHAVLAVG 311
Query 107 YGSEEGRDYWLAVNSWNEYWGDKGTFKIQMGE 138
YG E+ YWL NSW WGD G FK++MG+
Sbjct 312 YGVEDDVPYWLIKNSWGGEWGDNGYFKMEMGK 343
> hsa:1522 CTSZ, CTSX, FLJ17088; cathepsin Z (EC:3.4.18.1); K08568
cathepsin X [EC:3.4.18.1]
Length=303
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/131 (26%), Positives = 56/131 (42%), Gaps = 3/131 (2%)
Query 7 HHSDGPYPQCDGPLPKAPKCRK--DCEEVEYTTKVHPFKDDLHFASTSY-SVEGRDHIKK 63
H P C+ K +C K C + H ++ + Y S+ GR+ +
Sbjct 145 HQHGIPDETCNNYQAKDQECDKFNQCGTCNEFKECHAIRNYTLWRVGDYGSLSGREKMMA 204
Query 64 ELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWN 123
E+ NG ++ + E Y G+Y H V V G+G +G +YW+ NSW
Sbjct 205 EIYANGPISCGIMATERLANYTGGIYAEYQDTTYINHVVSVAGWGISDGTEYWIVRNSWG 264
Query 124 EYWGDKGTFKI 134
E WG++G +I
Sbjct 265 EPWGERGWLRI 275
> ath:AT4G23520 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=356
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 37/56 (66%), Gaps = 1/56 (1%)
Query 79 EDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGTFKI 134
++F++Y+ +Y+ G + HA+ ++GYGSE G+DYW+ NSW WGD G KI
Sbjct 274 QEFMLYRSCIYNGPCGTNLD-HALVIVGYGSENGQDYWIVRNSWGTTWGDAGYIKI 328
> pfa:PFB0360c SERA-1; serine repeat antigen 1 (SERA-1)
Length=994
Score = 58.9 bits (141), Expect = 7e-09, Method: Composition-based stats.
Identities = 32/84 (38%), Positives = 50/84 (59%), Gaps = 10/84 (11%)
Query 61 IKKELKENGTLTGAFLVFEDFLMYK---KGVYHHVTGIPMGGHAVKVIGYGS-----EEG 112
IK E+ NG++ A++ E+ L Y+ K V ++ G HAV ++GYG+ +E
Sbjct 673 IKDEIMNNGSVI-AYVKAENVLGYELNGKNV-QNLCGDKTPDHAVNIVGYGNYINDEDEK 730
Query 113 RDYWLAVNSWNEYWGDKGTFKIQM 136
+ YW+ NSW +YWGD+G FK+ M
Sbjct 731 KSYWIVRNSWGKYWGDEGYFKVDM 754
> pfa:PFB0335c SERA-6, SERP; serine repeat antigen 6 (SERA-6)
Length=1031
Score = 58.9 bits (141), Expect = 8e-09, Method: Composition-based stats.
Identities = 40/116 (34%), Positives = 60/116 (51%), Gaps = 16/116 (13%)
Query 35 YTTKVHPFKDDLHFAS--TSYSVEGRD----HIKKELKENGTLTGAFLVFEDFLMYK--- 85
+ KVH + + F S TSY D +K+E++ G++ ++ +D + Y
Sbjct 738 FNKKVHRYIGNKGFISHETSYFKNNMDLFIDMVKREVQNKGSVI-IYIKTQDVIGYDFNG 796
Query 86 KGVYHHVTGIPMGGHAVKVIGYGSE-----EGRDYWLAVNSWNEYWGDKGTFKIQM 136
KGV H + G HA +IGYG+ E R YWL NSW+ YWGD+G F++ M
Sbjct 797 KGV-HSMCGDRTPDHAANIIGYGNYINKKGEKRSYWLIRNSWSYYWGDEGNFRVDM 851
> xla:494800 ctsz; cathepsin Z (EC:3.4.18.1); K08568 cathepsin
X [EC:3.4.18.1]
Length=296
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 54 SVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEG 112
SV GR+ + E+ +NG ++ + E Y G+Y M H V V G+G E G
Sbjct 186 SVSGREKMMAEIYKNGPISCGIMATEKLDAYTGGLYAEFQPSAMINHIVSVAGWGLDENG 245
Query 113 RDYWLAVNSWNEYWGDKGTFKI 134
+YW+ NSW E WG++G +I
Sbjct 246 VEYWIVRNSWGEPWGERGWLRI 267
> ath:AT3G19400 cysteine proteinase, putative
Length=362
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 81 FLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGTFKIQ 135
F +YK GV GI + H V V+GYGS G DYW+ NSW WGD G K+Q
Sbjct 275 FQLYKSGVMTGTCGISLD-HGVVVVGYGSTSGEDYWIIRNSWGLNWGDSGYVKLQ 328
> cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin
C [EC:3.4.14.1]
Length=635
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 55/127 (43%), Gaps = 20/127 (15%)
Query 27 RKDCEEVEYTTKVHPFKDDLHFASTSYSVEGRDHIKKELKENGTLTGAFLVFEDFLMYKK 86
R CEE E + ++ + Y D +K+E+ +NG + A + L+Y
Sbjct 452 RIYCEEGE-----RMYAEEYGYVGGCYGCCDEDRMKEEIFKNGPIAVAMHIDTSLLVYDN 506
Query 87 GVYHHV---------------TGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGT 131
GVY + G HA+ ++G+G E G YW+ NSW WG KG
Sbjct 507 GVYDSIPNDHTKYCDLPNKQLNGWEYTNHAIAIVGWGEENGIPYWIIRNSWGANWGKKGY 566
Query 132 FKIQMGE 138
KI+ G+
Sbjct 567 AKIRRGK 573
> dre:324818 ctsh, fc44c02, wu:fc44c02, zgc:85774; cathepsin H
(EC:3.4.22.16); K01366 cathepsin H [EC:3.4.22.16]
Length=330
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 40/82 (48%), Gaps = 7/82 (8%)
Query 74 AFLVFEDFLMYKKGVY-----HHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGD 128
A+ V DF+ YK G+Y H+ T M HAV +GY E G YW+ NSW WG
Sbjct 247 AYEVTSDFMHYKDGIYTSTECHNTT--DMVNHAVLAVGYAEENGTPYWIVKNSWGTNWGI 304
Query 129 KGTFKIQMGEAGIDKEFCGGEP 150
KG F I+ G+ C P
Sbjct 305 KGYFYIERGKNMCGLAACSSYP 326
> pfa:PFB0330c SERA-7; serine repeat antigen 7 (SERA-7)
Length=946
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 50/84 (59%), Gaps = 10/84 (11%)
Query 61 IKKELKENGTLTGAFLVFE---DFLMYKKGVYHHVTGIPMGGHAVKVIGYGS---EEG-- 112
IK+E++ G++ A++ E DF KGV H++ G HA +IGYG+ EEG
Sbjct 678 IKREIQNKGSVI-AYIKTENVIDFDFNGKGV-HNMCGDKEPDHAANIIGYGNYIDEEGEK 735
Query 113 RDYWLAVNSWNEYWGDKGTFKIQM 136
+ YWL NSW YWGD+G F++ M
Sbjct 736 KSYWLIRNSWGYYWGDEGNFRVDM 759
> ath:AT3G19390 cysteine proteinase, putative / thiol protease,
putative
Length=452
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 81 FLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKGTFKIQ 135
F +Y GV+ G + H V +GYGSE G+DYW+ NSW WG+ G FK++
Sbjct 272 FQLYTSGVFTGTCGTSLD-HGVVAVGYGSEGGQDYWIVRNSWGSNWGESGYFKLE 325
> xla:432187 hypothetical protein MGC82409; K08568 cathepsin X
[EC:3.4.18.1]
Length=296
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 54 SVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEG 112
SV GR+ + E+ +NG ++ + + Y G+Y M H + V G+G E G
Sbjct 186 SVSGREKMMAEIYKNGPISCGIMATDKLDAYTGGLYAEYQPRAMINHIISVAGWGLDENG 245
Query 113 RDYWLAVNSWNEYWGDKGTFKI 134
+YW+ NSW E WG++G +I
Sbjct 246 VEYWIVRNSWGEPWGERGWLRI 267
> cel:M04G12.2 cpz-2; CathePsin Z family member (cpz-2); K08568
cathepsin X [EC:3.4.18.1]
Length=467
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 3/82 (3%)
Query 55 VEGRDHIKKELKENGTLTGAFLVFEDF-LMYKKGVYHHVTGIPMGGHAVKVIGYGSEE-G 112
V+GRD I E+K+ G + A + F Y KGVY + + H + + G+G +E G
Sbjct 354 VQGRDKIMSEIKKGGPIACAIGATKKFEYEYVKGVYSEKSDLE-SNHIISLTGWGVDENG 412
Query 113 RDYWLAVNSWNEYWGDKGTFKI 134
+YW+A NSW E WG+ G F++
Sbjct 413 VEYWIARNSWGEAWGELGWFRV 434
> hsa:1512 CTSH, ACC-4, ACC-5, CPSB, DKFZp686B24257, MGC1519,
minichain; cathepsin H (EC:3.4.22.16); K01366 cathepsin H [EC:3.4.22.16]
Length=335
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 39/80 (48%), Gaps = 3/80 (3%)
Query 74 AFLVFEDFLMYKKGVYHHVTGIPM---GGHAVKVIGYGSEEGRDYWLAVNSWNEYWGDKG 130
AF V +DF+MY+ G+Y + HAV +GYG + G YW+ NSW WG G
Sbjct 252 AFEVTQDFMMYRTGIYSSTSCHKTPDKVNHAVLAVGYGEKNGIPYWIVKNSWGPQWGMNG 311
Query 131 TFKIQMGEAGIDKEFCGGEP 150
F I+ G+ C P
Sbjct 312 YFLIERGKNMCGLAACASYP 331
> xla:380516 ctss-a, MGC69026; cathepsin S (EC:3.4.22.27); K01368
cathepsin S [EC:3.4.22.27]
Length=333
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Query 59 DHIKKELKENGTLTGAFL-VFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYGSEEGRDYWL 117
D++K+ L G ++ A F +YK GVY + H V IGYG+ G+D+WL
Sbjct 238 DNLKQALGTIGPISVAIDGTRPTFFLYKSGVYSDPSCSQEVNHGVLAIGYGTLNGQDFWL 297
Query 118 AVNSWNEYWGDKGTFKIQMGEAGI 141
NSW Y+GDKG +I + +
Sbjct 298 LKNSWGTYYGDKGFVRIARNKGNL 321
> mmu:64138 Ctsz, AI787083, AU019819, CTSX, D2Wsu143e; cathepsin
Z (EC:3.4.18.1); K08568 cathepsin X [EC:3.4.18.1]
Length=306
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 54 SVEGRDHIKKELKENGTLTGAFLVFEDFLMYKKGVYHHVTGIPMGGHAVKVIGYG-SEEG 112
S+ GR+ + E+ NG ++ + E Y G+Y + H + V G+G S +G
Sbjct 197 SLSGREKMMAEIYANGPISCGIMATEMMSNYTGGIYAEHQDQAVINHIISVAGWGVSNDG 256
Query 113 RDYWLAVNSWNEYWGDKGTFKI 134
+YW+ NSW E WG+KG +I
Sbjct 257 IEYWIVRNSWGEPWGEKGWMRI 278
Lambda K H
0.318 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40