bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1113_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
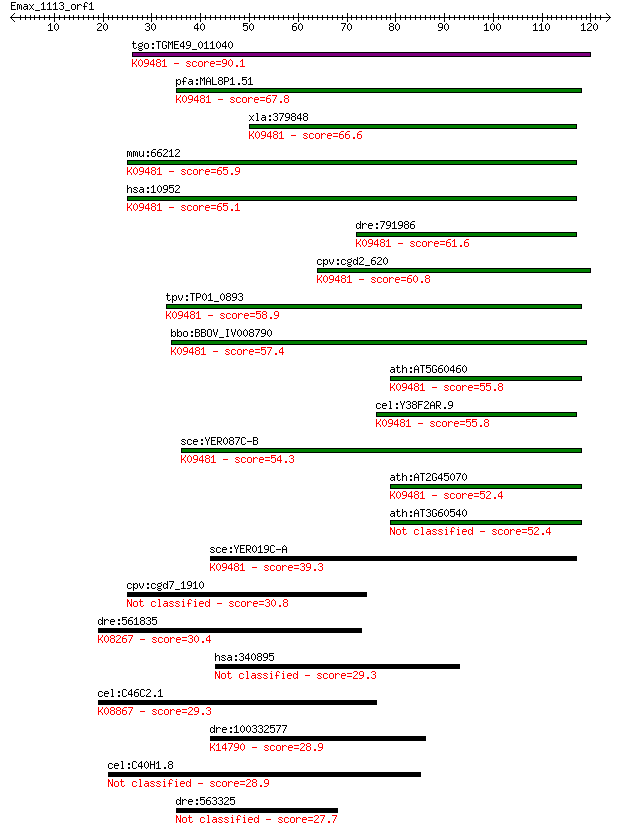
tgo:TGME49_011040 Sec61beta family protein ; K09481 protein tr... 90.1 2e-18
pfa:MAL8P1.51 Sec61 beta subunit, putative; K09481 protein tra... 67.8 8e-12
xla:379848 sec61b, MGC130844, MGC52846, sec61beta; Sec61 beta ... 66.6 2e-11
mmu:66212 Sec61b, 1190006C12Rik, AI326121, AW122942, MGC102034... 65.9 3e-11
hsa:10952 SEC61B; Sec61 beta subunit; K09481 protein transport... 65.1 5e-11
dre:791986 sec61b, MGC191409, MGC92922, fa20b05, wu:fa20b05, z... 61.6 5e-10
cpv:cgd2_620 hypothetical protein ; K09481 protein transport p... 60.8 9e-10
tpv:TP01_0893 hypothetical protein; K09481 protein transport p... 58.9 4e-09
bbo:BBOV_IV008790 23.m06502; hypothetical protein; K09481 prot... 57.4 1e-08
ath:AT5G60460 sec61beta family protein; K09481 protein transpo... 55.8 3e-08
cel:Y38F2AR.9 hypothetical protein; K09481 protein transport p... 55.8 3e-08
sce:YER087C-B SBH1, SEB1, YER087C-A; Beta subunit of the Sec61... 54.3 8e-08
ath:AT2G45070 SEC61 BETA; protein transporter; K09481 protein ... 52.4 3e-07
ath:AT3G60540 sec61beta family protein 52.4 3e-07
sce:YER019C-A SBH2, SEB2; Sbh2p; K09481 protein transport prot... 39.3 0.003
cpv:cgd7_1910 coatomer SEC21 gamma subunit like (beta adaptin) 30.8 1.2
dre:561835 rictor; RPTOR independent companion of MTOR, comple... 30.4 1.3
hsa:340895 C10orf112, FLJ60310, bA265G8.2; chromosome 10 open ... 29.3 2.8
cel:C46C2.1 wnk-1; mammalian WNK-type protein kinase homolog f... 29.3 3.4
dre:100332577 hypothetical protein LOC100332577; K14790 nucleo... 28.9 3.6
cel:C40H1.8 hypothetical protein 28.9 4.1
dre:563325 fa04d06, wu:fa04d06; si:dkey-92i17.2 27.7 8.1
> tgo:TGME49_011040 Sec61beta family protein ; K09481 protein
transport protein SEC61 subunit beta
Length=99
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 50/96 (52%), Positives = 66/96 (68%), Gaps = 5/96 (5%)
Query 26 MVGTNSLSTQASQTT-VGGSRVAATRKRVTSKSSTASSGAATISRPRS-NAANQGILKFY 83
MVGTN+ S QAS VGG+R + R+RVT+ + +A R R A++QGIL+FY
Sbjct 1 MVGTNAPSAQASSAQHVGGARTSVPRRRVTAGGGAS---SAAGQRSRGVPASSQGILRFY 57
Query 84 TEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKIRQ 119
T+D PGL++GPQ VLI+ L FM VV+LHIVGK+ Q
Sbjct 58 TDDTPGLKIGPQTVLILTLCFMASVVLLHIVGKVHQ 93
> pfa:MAL8P1.51 Sec61 beta subunit, putative; K09481 protein transport
protein SEC61 subunit beta
Length=80
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 51/83 (61%), Gaps = 4/83 (4%)
Query 35 QASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSNAANQGILKFYTEDAPGLRVGP 94
A+ VGG R A R+ +S+ +++G + R SN + I+KFY +D+PG ++ P
Sbjct 2 NAAPVIVGGMRTPARRR----QSNASANGNNSKQRRNSNGNSNSIVKFYGDDSPGFKLTP 57
Query 95 QAVLIMALVFMGVVVMLHIVGKI 117
Q VLI L+FM VV+LHI+ KI
Sbjct 58 QTVLISTLIFMASVVILHIISKI 80
> xla:379848 sec61b, MGC130844, MGC52846, sec61beta; Sec61 beta
subunit; K09481 protein transport protein SEC61 subunit beta
Length=96
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 45/67 (67%), Gaps = 6/67 (8%)
Query 50 RKRVTSKSSTASSGAATISRPRSNAANQGILKFYTEDAPGLRVGPQAVLIMALVFMGVVV 109
R+R + SST S+G T +A G+ +FYTED+PGL+VGP VL+M+L+F+ V
Sbjct 32 RQRKNASSSTRSAGRTT------SAGTGGMWRFYTEDSPGLKVGPVPVLVMSLLFIASVF 85
Query 110 MLHIVGK 116
MLHI GK
Sbjct 86 MLHIWGK 92
> mmu:66212 Sec61b, 1190006C12Rik, AI326121, AW122942, MGC102034;
Sec61 beta subunit; K09481 protein transport protein SEC61
subunit beta
Length=96
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 8/94 (8%)
Query 25 TMVGTNSLSTQASQTTVGGSRVAAT--RKRVTSKSSTASSGAATISRPRSNAANQGILKF 82
T GTN S+ S + +R A + R+R + T S+G T +A G+ +F
Sbjct 5 TPSGTNVGSSGRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTT------SAGTGGMWRF 58
Query 83 YTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
YTED+PGL+VGP VL+M+L+F+ V MLHI GK
Sbjct 59 YTEDSPGLKVGPVPVLVMSLLFIAAVFMLHIWGK 92
> hsa:10952 SEC61B; Sec61 beta subunit; K09481 protein transport
protein SEC61 subunit beta
Length=96
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 55/94 (58%), Gaps = 8/94 (8%)
Query 25 TMVGTNSLSTQASQTTVGGSRVAAT--RKRVTSKSSTASSGAATISRPRSNAANQGILKF 82
T GTN S+ S + +R A + R+R + T S+G T +A G+ +F
Sbjct 5 TPSGTNVGSSGRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTT------SAGTGGMWRF 58
Query 83 YTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
YTED+PGL+VGP VL+M+L+F+ V MLHI GK
Sbjct 59 YTEDSPGLKVGPVPVLVMSLLFIASVFMLHIWGK 92
> dre:791986 sec61b, MGC191409, MGC92922, fa20b05, wu:fa20b05,
zgc:92922; Sec61 beta subunit; K09481 protein transport protein
SEC61 subunit beta
Length=97
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 25/45 (55%), Positives = 35/45 (77%), Gaps = 0/45 (0%)
Query 72 SNAANQGILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
++A G+ +FYTED+PGL+VGP VL+M+L+F+ V MLHI GK
Sbjct 49 ASAGTGGMWRFYTEDSPGLKVGPVPVLVMSLLFIASVFMLHIWGK 93
> cpv:cgd2_620 hypothetical protein ; K09481 protein transport
protein SEC61 subunit beta
Length=66
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 41/56 (73%), Gaps = 1/56 (1%)
Query 64 AATISRPRSNAANQGILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKIRQ 119
A++++ R N A Q ++ +Y +D PGL++GP VL+M L +M +V++LHI+GK ++
Sbjct 8 ASSVTSSRENLA-QNLMSYYVDDTPGLKLGPMTVLVMTLAYMSIVIVLHILGKFKE 62
> tpv:TP01_0893 hypothetical protein; K09481 protein transport
protein SEC61 subunit beta
Length=86
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 55/86 (63%), Gaps = 11/86 (12%)
Query 33 STQASQTT-VGGSRVAATRKRVTSKSSTASSGAATISRPRSNAANQGILKFYTEDAPGLR 91
S Q +Q+T VGG RV+ TR+R TS+S+ G T P N G K+Y + GL
Sbjct 5 SKQPAQSTMVGGQRVS-TRRRTTSEST----GTRTNLPP-----NSGFQKYYGLTSTGLE 54
Query 92 VGPQAVLIMALVFMGVVVMLHIVGKI 117
+ Q+VL++AL++MGVVV++HIV +I
Sbjct 55 MSQQSVLLLALIYMGVVVVMHIVSRI 80
> bbo:BBOV_IV008790 23.m06502; hypothetical protein; K09481 protein
transport protein SEC61 subunit beta
Length=84
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 10/86 (11%)
Query 34 TQASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSN-AANQGILKFYTEDAPGLRV 92
T A T +GG R+A RKR S+++TA + R+N N G +Y +++
Sbjct 5 TPAQSTMIGGQRMAP-RKRAHSETNTAGT--------RTNLPPNSGFQHYYGFATSSIQL 55
Query 93 GPQAVLIMALVFMGVVVMLHIVGKIR 118
GPQAVL++A+ +MG VV++HIV +I+
Sbjct 56 GPQAVLLLAVCYMGAVVVMHIVSRIK 81
> ath:AT5G60460 sec61beta family protein; K09481 protein transport
protein SEC61 subunit beta
Length=109
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 22/39 (56%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM+L F+G V LH+ GK+
Sbjct 62 MLRFYTDDAPGLKISPTVVLIMSLCFIGFVTALHVFGKL 100
> cel:Y38F2AR.9 hypothetical protein; K09481 protein transport
protein SEC61 subunit beta
Length=81
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 23/41 (56%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 76 NQGILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGK 116
N G+ +FYTED+ GL++GP VL+M+LVF+ V +LHI GK
Sbjct 35 NGGLWRFYTEDSTGLKIGPVPVLVMSLVFIASVFVLHIWGK 75
> sce:YER087C-B SBH1, SEB1, YER087C-A; Beta subunit of the Sec61p
ER translocation complex (Sec61p-Sss1p-Sbh1p); involved
in protein translocation into the endoplasmic reticulum; interacts
with the exocyst complex and also with Rtn1p; homologous
to Sbh2p; K09481 protein transport protein SEC61 subunit
beta
Length=82
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 48/83 (57%), Gaps = 8/83 (9%)
Query 36 ASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSNA-ANQGILKFYTEDAPGLRVGP 94
+S T GG R RK+ +S+ AS+ P+ N +N ILK Y+++A GLRV P
Sbjct 2 SSPTPPGGQRTLQKRKQGSSQKVAASA-------PKKNTNSNNSILKIYSDEATGLRVDP 54
Query 95 QAVLIMALVFMGVVVMLHIVGKI 117
VL +A+ F+ VV LH++ K+
Sbjct 55 LVVLFLAVGFIFSVVALHVISKV 77
> ath:AT2G45070 SEC61 BETA; protein transporter; K09481 protein
transport protein SEC61 subunit beta
Length=82
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 32/39 (82%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM++ F+ V +LH++GK+
Sbjct 40 MLQFYTDDAPGLKISPNVVLIMSIGFIAFVAVLHVMGKL 78
> ath:AT3G60540 sec61beta family protein
Length=81
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 32/39 (82%), Gaps = 0/39 (0%)
Query 79 ILKFYTEDAPGLRVGPQAVLIMALVFMGVVVMLHIVGKI 117
+L+FYT+DAPGL++ P VLIM++ F+ V +LH++GK+
Sbjct 37 MLQFYTDDAPGLKISPNVVLIMSIGFIAFVAVLHVMGKL 75
> sce:YER019C-A SBH2, SEB2; Sbh2p; K09481 protein transport protein
SEC61 subunit beta
Length=88
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 42 GGSRVAATRKRVTS-KSSTASSGAATISRPRSNAANQGILKFYTEDAPGLRVGPQAVLIM 100
GG R+ R++ S K A + + ++ ILK YT++A G RV VL +
Sbjct 8 GGQRILQKRRQAQSIKEKQAKQTPTSTRQAGYGGSSSSILKLYTDEANGFRVDSLVVLFL 67
Query 101 ALVFMGVVVMLHIVGK 116
++ F+ V+ LH++ K
Sbjct 68 SVGFIFSVIALHLLTK 83
> cpv:cgd7_1910 coatomer SEC21 gamma subunit like (beta adaptin)
Length=936
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 28/49 (57%), Gaps = 5/49 (10%)
Query 25 TMVGTNSLSTQASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSN 73
T++ NSL T ++TV GS++ + +TS S AS A ISR R N
Sbjct 608 TLLDLNSLPTDPVESTVNGSQITSN---ITSVS--ASMEALNISRTRGN 651
> dre:561835 rictor; RPTOR independent companion of MTOR, complex
2; K08267 rapamycin-insensitive companion of mTOR
Length=1729
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 32/72 (44%), Gaps = 18/72 (25%)
Query 19 SFTLLFTMVGTNSLSTQASQTTVGGSRV------------------AATRKRVTSKSSTA 60
S +L + VGT +L TQ S +++G V +R+R+ ST+
Sbjct 1169 SVSLETSFVGTKTLDTQDSTSSIGEQEVRLNRTAERSSGVGDTHREQTSRERLAGDGSTS 1228
Query 61 SSGAATISRPRS 72
SGA SR +S
Sbjct 1229 GSGAQFKSRSQS 1240
> hsa:340895 C10orf112, FLJ60310, bA265G8.2; chromosome 10 open
reading frame 112
Length=1542
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 27/56 (48%), Gaps = 10/56 (17%)
Query 43 GSRVAATRKRVTSKSSTASSGAATIS------RPRSNAANQGILKFYTEDAPGLRV 92
GS+ + + TSKS AS G T+ PRS GILK YT + GL +
Sbjct 1221 GSKEGSVARITTSKSFPASLGMCTVRFWFYMIDPRS----MGILKVYTIEESGLNI 1272
> cel:C46C2.1 wnk-1; mammalian WNK-type protein kinase homolog
family member (wnk-1); K08867 WNK lysine deficient protein
kinase [EC:2.7.11.1]
Length=1838
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 26/67 (38%), Positives = 33/67 (49%), Gaps = 11/67 (16%)
Query 19 SFTLLFTMVGTNSLSTQAS----------QTTVGGSRVAATRKRVTSKSSTASSGAATIS 68
+ T L + +G SLS AS TTV S V ATR + S+SST SS + S
Sbjct 1705 TLTALQSALGNASLSLPASPPPNTETTKVNTTVIPSDVLATRMTM-SQSSTKSSNVSVSS 1763
Query 69 RPRSNAA 75
R R N +
Sbjct 1764 RHRDNQS 1770
> dre:100332577 hypothetical protein LOC100332577; K14790 nucleolar
protein 9
Length=604
Score = 28.9 bits (63), Expect = 3.6, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 20/44 (45%), Gaps = 0/44 (0%)
Query 42 GGSRVAATRKRVTSKSSTASSGAATISRPRSNAANQGILKFYTE 85
GG R + +R + ASSGA + R R + G + TE
Sbjct 9 GGQRSSKNDQRARKRPQQASSGAGQLKRERLDGTTVGYFRRVTE 52
> cel:C40H1.8 hypothetical protein
Length=342
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 28/64 (43%), Gaps = 0/64 (0%)
Query 21 TLLFTMVGTNSLSTQASQTTVGGSRVAATRKRVTSKSSTASSGAATISRPRSNAANQGIL 80
T+ + T + ST+ S T G T+++V S S + G I + G+L
Sbjct 91 TMKLYQIRTVNCSTKYSDVTCVGYTAFDTKRKVISISFKGAHGQDQIEEMMEDGMKYGLL 150
Query 81 KFYT 84
+YT
Sbjct 151 PYYT 154
> dre:563325 fa04d06, wu:fa04d06; si:dkey-92i17.2
Length=389
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 35 QASQTTVGGSRVAATRKRVTSKSSTASSGAATI 67
Q S++ V RKRV SK+ ++SSGAA +
Sbjct 124 QPSESLTSDRAVLPARKRVQSKNLSSSSGAAGV 156
Lambda K H
0.324 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40