bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1135_orf1
Length=215
Score E
Sequences producing significant alignments: (Bits) Value
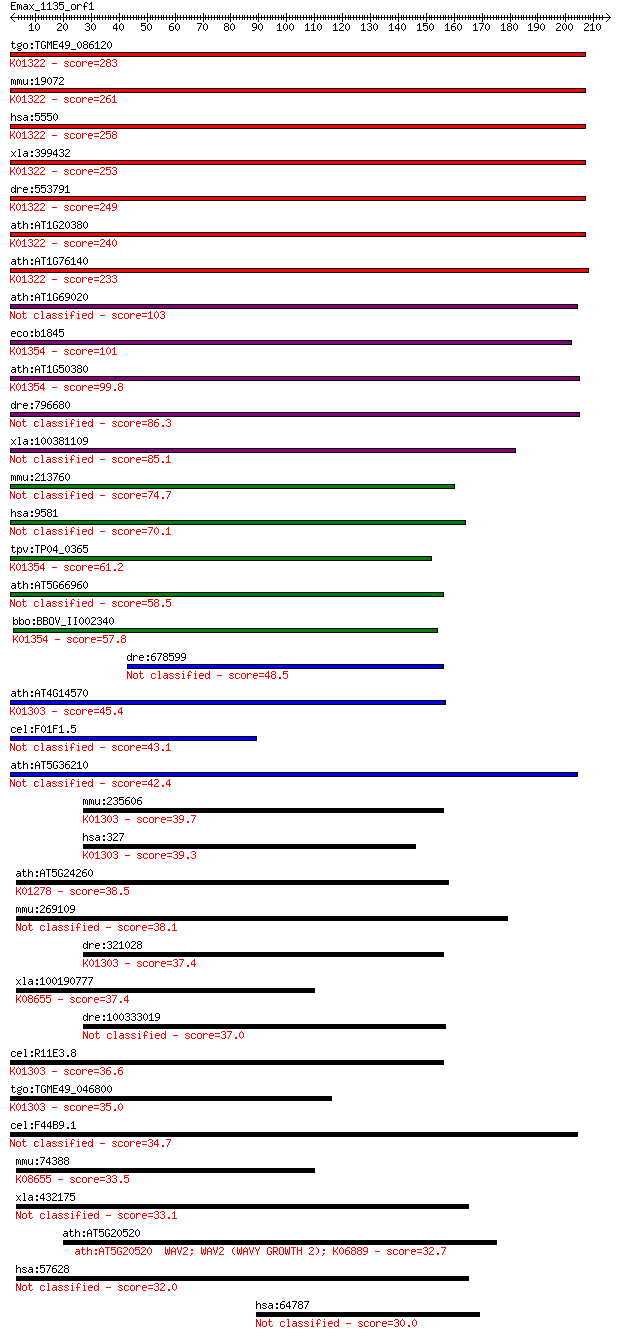
tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26... 283 4e-76
mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prol... 261 2e-69
hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4... 258 9e-69
xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26); K... 253 3e-67
dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl end... 249 7e-66
ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopep... 240 3e-63
ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase... 233 5e-61
ath:AT1G69020 prolyl oligopeptidase family protein 103 4e-22
eco:b1845 ptrB, ECK1846, JW1834, opdB, tlp; protease II (EC:3.... 101 2e-21
ath:AT1G50380 prolyl oligopeptidase family protein; K01354 oli... 99.8 6e-21
dre:796680 prepl, si:dkey-202m23.1; prolyl endopeptidase-like 86.3 7e-17
xla:100381109 prepl; prolyl endopeptidase-like (EC:3.4.21.-) 85.1 2e-16
mmu:213760 Prepl, 2810457N15Rik, 9530014L06Rik, D030028O16Rik,... 74.7 2e-13
hsa:9581 PREPL, FLJ16627, KIAA0436; prolyl endopeptidase-like ... 70.1 5e-12
tpv:TP04_0365 protease II (EC:3.4.21.83); K01354 oligopeptidas... 61.2 3e-09
ath:AT5G66960 prolyl oligopeptidase family protein 58.5 1e-08
bbo:BBOV_II002340 18.m06190; hypothetical protein; K01354 olig... 57.8 3e-08
dre:678599 MGC136971, si:dkey-16c7.3, wu:fc20g05; zgc:136971 48.5 2e-05
ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylamin... 45.4 1e-04
cel:F01F1.5 dpf-4; Dipeptidyl Peptidase Four (IV) family membe... 43.1 7e-04
ath:AT5G36210 serine-type peptidase 42.4 0.001
mmu:235606 Apeh, MGC38101; acylpeptide hydrolase (EC:3.4.19.1)... 39.7 0.009
hsa:327 APEH, ACPH, APH, D3F15S2, D3S48E, DNF15S2, MGC2178, OP... 39.3 0.010
ath:AT5G24260 prolyl oligopeptidase family protein; K01278 dip... 38.5 0.019
mmu:269109 Dpp10, 6430601K09Rik, DPP_X, Dprp3; dipeptidylpepti... 38.1 0.026
dre:321028 apeh, cb5, sb:cb5, wu:fi37d02; acylpeptide hydrolas... 37.4 0.035
xla:100190777 dpp8-a, dp8, dpp8, dpp8a, dprp1; dipeptidyl-pept... 37.4 0.040
dre:100333019 N-acylaminoacyl-peptide hydrolase-like 37.0 0.048
cel:R11E3.8 dpf-5; Dipeptidyl Peptidase Four (IV) family membe... 36.6 0.070
tgo:TGME49_046800 acylamino-acid-releasing enzyme, putative (E... 35.0 0.21
cel:F44B9.1 dpf-6; Dipeptidyl Peptidase Four (IV) family membe... 34.7 0.27
mmu:74388 Dpp8, 2310004I03Rik, 4932434F09Rik, AI666706, DPP_VI... 33.5 0.53
xla:432175 dpp10, MGC84485; dipeptidyl-peptidase 10 (non-funct... 33.1 0.78
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 32.7 1.1
hsa:57628 DPP10, DPL2, DPPY, DPRP3; dipeptidyl-peptidase 10 (n... 32.0 1.7
hsa:64787 EPS8L2, EPS8R2, FLJ16738, FLJ21935, FLJ22171, MGC126... 30.0 5.9
> tgo:TGME49_086120 prolyl endopeptidase, putative (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=825
Score = 283 bits (723), Expect = 4e-76, Method: Compositional matrix adjust.
Identities = 133/208 (63%), Positives = 159/208 (76%), Gaps = 2/208 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG WY+AAIK KQVSYDDFQ AAEYL + Y++P QLAI GGSNGGLLVGA
Sbjct 615 NIRGGGEYGRAWYEAAIKTKKQVSYDDFQQAAEYLIAQKYTSPQQLAIEGGSNGGLLVGA 674
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI-- 118
C NQRPDL+ A + VGV+DLLRF+KFTIGHAW SDYGNP+ EEDF I ISP+HNI
Sbjct 675 CINQRPDLYGAAIIHVGVLDLLRFHKFTIGHAWTSDYGNPENEEDFPAILAISPLHNIGK 734
Query 119 NPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGH 178
+ + PAVL++T DHDDRVSPFHSLKYIA+LQH+VG S + PL+ +DT GH
Sbjct 735 GRGKGKGHQYPAVLLLTGDHDDRVSPFHSLKYIAELQHSVGSSPKQTNPLVIRVDTNTGH 794
Query 179 GGGKPLMKVLDEQALTYGFIASVLGLKW 206
G GKP+ K ++E A YGF+A+ L ++W
Sbjct 795 GAGKPVKKTIEEAADVYGFLANALHIQW 822
> mmu:19072 Prep, AI047692, AI450383, D10Wsu136e, PEP, Pop; prolyl
endopeptidase (EC:3.4.21.26); K01322 prolyl oligopeptidase
[EC:3.4.21.26]
Length=710
Score = 261 bits (666), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 119/206 (57%), Positives = 153/206 (74%), Gaps = 0/206 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K I NKQ +DDFQ AAEYL +GY++P +L I GGSNGGLLV A
Sbjct 503 NIRGGGEYGETWHKGGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAA 562
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
CANQRPDLF V+A+VGVMD+L+F+KFTIGHAW +DYG D ++ F+++ K SP+HN+
Sbjct 563 CANQRPDLFGCVIAQVGVMDMLKFHKFTIGHAWTTDYGCSDTKQHFEWLLKYSPLHNVKL 622
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ P++L++TADHDDRV P HSLK+IA LQ+ VG S S PL+ H+DTKAGHG
Sbjct 623 PEADDIQYPSMLLLTADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGA 682
Query 181 GKPLMKVLDEQALTYGFIASVLGLKW 206
GKP KV++E + + FIA L ++W
Sbjct 683 GKPTAKVIEEVSDMFAFIARCLNIEW 708
> hsa:5550 PREP, MGC16060, PE, PEP; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=710
Score = 258 bits (660), Expect = 9e-69, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 152/206 (73%), Gaps = 0/206 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K I NKQ +DDFQ AAEYL +GY++P +L I GGSNGGLLV A
Sbjct 503 NIRGGGEYGETWHKGGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAA 562
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
CANQRPDLF V+A+VGVMD+L+F+K+TIGHAW +DYG D ++ F+++ K SP+HN+
Sbjct 563 CANQRPDLFGCVIAQVGVMDMLKFHKYTIGHAWTTDYGCSDSKQHFEWLVKYSPLHNVKL 622
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ P++L++TADHDDRV P HSLK+IA LQ+ VG S S PL+ H+DTKAGHG
Sbjct 623 PEADDIQYPSMLLLTADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGA 682
Query 181 GKPLMKVLDEQALTYGFIASVLGLKW 206
GKP KV++E + + FIA L + W
Sbjct 683 GKPTAKVIEEVSDMFAFIARCLNVDW 708
> xla:399432 prep, xprep; prolyl endopeptidase (EC:3.4.21.26);
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=712
Score = 253 bits (646), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 149/206 (72%), Gaps = 0/206 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+KA NKQ +DDFQ AAEYL +GYS+ + I GGSNGGLLV A
Sbjct 505 NIRGGGEYGETWHKAGSLGNKQNCFDDFQCAAEYLVKEGYSSAKNITINGGSNGGLLVAA 564
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
C NQRPDLF +A+VGVMD+L+F+KFTIGHAW +D+G D +E F ++ K SP+HNI
Sbjct 565 CTNQRPDLFGCTIAQVGVMDMLKFHKFTIGHAWTTDFGCSDNKEHFDWLIKYSPLHNIRV 624
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ P++L++TADHDDRV P HSLK+IA LQ+ VG S + PL+ H+DTKAGHG
Sbjct 625 PEKDGIQYPSMLLLTADHDDRVVPLHSLKFIASLQNIVGRSPNQTNPLLIHVDTKAGHGA 684
Query 181 GKPLMKVLDEQALTYGFIASVLGLKW 206
GKP KV++E + + FIA LGL+W
Sbjct 685 GKPTAKVIEEVSDMFAFIAQCLGLQW 710
> dre:553791 prep, MGC110670, im:7140031, zgc:110670; prolyl endopeptidase
(EC:3.4.21.26); K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=709
Score = 249 bits (635), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 144/206 (69%), Gaps = 0/206 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K + NKQ + DFQ AAEYL +GY++P +L I GGSNGGLLV A
Sbjct 502 NIRGGGEYGETWHKGGMLANKQNCFTDFQCAAEYLIKEGYTSPKKLTINGGSNGGLLVAA 561
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINP 120
C NQRPDLF VA+VGVMD+L+F+KFTIGHAW +D+G + +E F ++ K SP+HNI
Sbjct 562 CVNQRPDLFGCAVAQVGVMDMLKFHKFTIGHAWTTDFGCSEIKEQFDWLIKYSPLHNIQV 621
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAGHGG 180
+ PAVL++T DHDDRV P HSLKYIA LQ+ +G PL +IDTK+GHG
Sbjct 622 PEGDGVQYPAVLLLTGDHDDRVVPLHSLKYIATLQNVIGQCPGQKNPLFIYIDTKSGHGA 681
Query 181 GKPLMKVLDEQALTYGFIASVLGLKW 206
GKP KV+ E A TY FIA L L W
Sbjct 682 GKPTSKVIQEVADTYAFIARCLNLSW 707
> ath:AT1G20380 prolyl oligopeptidase, putative / prolyl endopeptidase,
putative / post-proline cleaving enzyme, putative;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=731
Score = 240 bits (612), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 114/213 (53%), Positives = 151/213 (70%), Gaps = 7/213 (3%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+K+ NKQ +DDF S AEYL + GY+ P +L I GGSNGG+LVGA
Sbjct 517 NIRGGGEYGEEWHKSGALANKQNCFDDFISGAEYLVSAGYTQPRKLCIEGGSNGGILVGA 576
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI-N 119
C NQRPDLF +A VGVMD+LRF+KFTIGHAW S++G DKEE+F ++ K SP+HN+
Sbjct 577 CINQRPDLFGCALAHVGVMDMLRFHKFTIGHAWTSEFGCSDKEEEFHWLIKYSPLHNVKR 636
Query 120 PEVVRSD---KQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYS---SPVSTPLIAHID 173
P ++D + P+ +++TADHDDRV P HS K +A +Q+ +G S SP + P+IA I+
Sbjct 637 PWEQKTDLFFQYPSTMLLTADHDDRVVPLHSYKLLATMQYELGLSLENSPQTNPIIARIE 696
Query 174 TKAGHGGGKPLMKVLDEQALTYGFIASVLGLKW 206
KAGHG G+P K++DE A Y F+A ++ W
Sbjct 697 VKAGHGAGRPTQKMIDEAADRYSFMAKMVDASW 729
> ath:AT1G76140 serine-type endopeptidase/ serine-type peptidase;
K01322 prolyl oligopeptidase [EC:3.4.21.26]
Length=792
Score = 233 bits (593), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 111/211 (52%), Positives = 145/211 (68%), Gaps = 4/211 (1%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
NIRGGGEYG W+KA KQ +DDF S AEYL + GY+ P +L I GGSNGGLLVGA
Sbjct 581 NIRGGGEYGEEWHKAGSLAKKQNCFDDFISGAEYLVSAGYTQPSKLCIEGGSNGGLLVGA 640
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI-N 119
C NQRPDL+ +A VGVMD+LRF+KFTIGHAW SDYG + EE+F ++ K SP+HN+
Sbjct 641 CINQRPDLYGCALAHVGVMDMLRFHKFTIGHAWTSDYGCSENEEEFHWLIKYSPLHNVKR 700
Query 120 PEVVRSD---KQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKA 176
P ++D + P+ +++TADHDDRV P HSLK +A + +SP P+I I+ KA
Sbjct 701 PWEQQTDHLVQYPSTMLLTADHDDRVVPLHSLKLLAHVLCTSLDNSPQMNPIIGRIEVKA 760
Query 177 GHGGGKPLMKVLDEQALTYGFIASVLGLKWS 207
GHG G+P K++DE A Y F+A ++ W+
Sbjct 761 GHGAGRPTQKMIDEAADRYSFMAKMVNASWT 791
> ath:AT1G69020 prolyl oligopeptidase family protein
Length=757
Score = 103 bits (257), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 67/208 (32%), Positives = 104/208 (50%), Gaps = 13/208 (6%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGG W+K+ + KQ S DF +A+YL KGY LA +G S G +L A
Sbjct 553 DVRGGGSGEFSWHKSGTRSLKQNSIQDFIYSAKYLVEKGYVHRHHLAAVGYSAGAILPAA 612
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVS-----DYGNPDKEEDFKYIYKISPI 115
N P LF+AV+ KV +D+L N + + ++ ++GNPD + DF I SP
Sbjct 613 AMNMHPSLFQAVILKVPFVDVL--NTLSDPNLPLTLLDHEEFGNPDNQTDFGSILSYSPY 670
Query 116 HNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTK 175
I +V P++LV T+ HD RV + K++A+++ + + S +I +
Sbjct 671 DKIRKDVC----YPSMLVTTSFHDSRVGVWEGAKWVAKIRDSTCHD--CSRAVILKTNMN 724
Query 176 AGHGGGKPLMKVLDEQALTYGFIASVLG 203
GH G +E A Y F+ V+G
Sbjct 725 GGHFGEGGRYAQCEETAFDYAFLLKVMG 752
> eco:b1845 ptrB, ECK1846, JW1834, opdB, tlp; protease II (EC:3.4.21.83);
K01354 oligopeptidase B [EC:3.4.21.83]
Length=686
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/204 (29%), Positives = 101/204 (49%), Gaps = 12/204 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G WY+ K+ +++D+ A + L GY +P MGGS GG+L+G
Sbjct 481 HVRGGGELGQQWYEDGKFLKKKNTFNDYLDACDALLKLGYGSPSLCYAMGGSAGGMLMGV 540
Query 61 CANQRPDLFRAVVAKVGVMDLLRF---NKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHN 117
NQRP+LF V+A+V +D++ + ++GNP + ++Y+ SP N
Sbjct 541 AINQRPELFHGVIAQVPFVDVVTTMLDESIPLTTGEFEEWGNPQDPQYYEYMKSYSPYDN 600
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAG 177
+ + P +LV T HD +V + K++A+L+ L+ D +G
Sbjct 601 VTAQAY-----PHLLVTTGLHDSQVQYWEPAKWVAKLREL----KTDDHLLLLCTDMDSG 651
Query 178 HGGGKPLMKVLDEQALTYGFIASV 201
HGG K + A+ Y F+ ++
Sbjct 652 HGGKSGRFKSYEGVAMEYAFLVAL 675
> ath:AT1G50380 prolyl oligopeptidase family protein; K01354 oligopeptidase
B [EC:3.4.21.83]
Length=710
Score = 99.8 bits (247), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 64/207 (30%), Positives = 103/207 (49%), Gaps = 12/207 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G WY+ K+ ++ DF + AE L Y + ++L + G S GGLL+GA
Sbjct 508 HVRGGGEMGRQWYENGKLLKKKNTFTDFIACAERLIELKYCSKEKLCMEGRSAGGLLMGA 567
Query 61 CANQRPDLFRAVVAKVGVMDLLRFN---KFTIGHAWVSDYGNPDKEEDFKYIYKISPIHN 117
N RPDLF+ V+A V +D+L + + ++G+P KEE + Y+ SP+ N
Sbjct 568 VVNMRPDLFKVVIAGVPFVDVLTTMLDPTIPLTTSEWEEWGDPRKEEFYFYMKSYSPVDN 627
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAG 177
+ + P +LV +D RV K++A+L+ + L+ + AG
Sbjct 628 VTAQ-----NYPNMLVTAGLNDPRVMYSEPGKWVAKLREM----KTDNNVLLFKCELGAG 678
Query 178 HGGGKPLMKVLDEQALTYGFIASVLGL 204
H + L E A T+ F+ VL +
Sbjct 679 HFSKSGRFEKLQEDAFTFAFMMKVLDM 705
> dre:796680 prepl, si:dkey-202m23.1; prolyl endopeptidase-like
Length=692
Score = 86.3 bits (212), Expect = 7e-17, Method: Composition-based stats.
Identities = 60/213 (28%), Positives = 97/213 (45%), Gaps = 14/213 (6%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G+ W++A KQ +D + + L G S PD A+ S G +L GA
Sbjct 473 HVRGGGERGLSWHRAGSVLQKQRGVEDLAACIQTLHRLGVSGPDLTALTARSAGAILAGA 532
Query 61 CANQRPDLFRAVVAKVGVMDLL---RFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHN 117
NQ P L RA++ + +D+L + + ++G+P E I P HN
Sbjct 533 LCNQNPQLIRALILQAPFLDVLGTMQDTSLPLTLEEKGEWGDPLIREHRDNIASYCPCHN 592
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSP-----VSTP-LIAH 171
I P++ P++L+ D RV +K++ +L+ A+ + P +I
Sbjct 593 IQPQLY-----PSMLITAYSEDHRVPVSGVIKFVERLKKAIQMCTTQVDINARVPSVILD 647
Query 172 IDTKAGHGGGKPLMKVLDEQALTYGFIASVLGL 204
I A H G + L+E A F+ + LGL
Sbjct 648 IQPGADHFGPEDFHLSLNESARQLAFLYTELGL 680
> xla:100381109 prepl; prolyl endopeptidase-like (EC:3.4.21.-)
Length=707
Score = 85.1 bits (209), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 61/190 (32%), Positives = 85/190 (44%), Gaps = 16/190 (8%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G W+ + + K +D S +L GYS P A+ S GG+L GA
Sbjct 487 HVRGGGELGCNWHSEGVLDKKLNGLEDLGSCISHLHGLGYSQPHYSAVEAASAGGVLAGA 546
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNK-----FTIGHAWVSDYGNPDKEEDF-KYIYKISP 114
N P LFRAVV + +D+L TI ++GNP +E + +YI P
Sbjct 547 LCNSAPRLFRAVVLEAPFLDVLNTMMNVSLPLTIEEQ--EEWGNPLSDEKYHRYIKSYCP 604
Query 115 IHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAV---GYSSPVSTPLIAH 171
NI P+ P V + ++D RV L YI +L+ A + S S I H
Sbjct 605 YQNITPQ-----NYPCVRITAYENDQRVPIQGLLGYITRLRKAARDYCHESGTSESRIPH 659
Query 172 IDTKAGHGGG 181
I GG
Sbjct 660 IYLDVHPGGS 669
> mmu:213760 Prepl, 2810457N15Rik, 9530014L06Rik, D030028O16Rik,
MGC7980, mKIAA0436; prolyl endopeptidase-like (EC:3.4.21.-)
Length=725
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 48/163 (29%), Positives = 79/163 (48%), Gaps = 9/163 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G+ W+ K D + + L ++G+S P + S GG+LVGA
Sbjct 506 HVRGGGELGLQWHADGRLTKKLNGLADLVACIKTLHSQGFSQPSLTTLSAFSAGGVLVGA 565
Query 61 CANQRPDLFRAVVAKVGVMDLLRF---NKFTIGHAWVSDYGNPDKEEDFK-YIYKISPIH 116
N +P+L RAV + +D+L + + ++GNP +E K YI + P
Sbjct 566 LCNSKPELLRAVTLEAPFLDVLNTMLDTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPCQ 625
Query 117 NINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVG 159
NI P+ P+V + ++D+RV + Y +L+ AV
Sbjct 626 NIKPQ-----HYPSVHITAYENDERVPLKGIVNYTEKLKEAVA 663
> hsa:9581 PREPL, FLJ16627, KIAA0436; prolyl endopeptidase-like
(EC:3.4.21.-)
Length=665
Score = 70.1 bits (170), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 48/176 (27%), Positives = 81/176 (46%), Gaps = 18/176 (10%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G+ W+ K D ++ + L +G+S P + S GG+L GA
Sbjct 446 HVRGGGELGLQWHADGRLTKKLNGLADLEACIKTLHGQGFSQPSLTTLTAFSAGGVLAGA 505
Query 61 CANQRPDLFRAVVAKVGVMDLLRF---NKFTIGHAWVSDYGNPDKEEDFK-YIYKISPIH 116
N P+L RAV + +D+L + + ++GNP +E K YI + P
Sbjct 506 LCNSNPELVRAVTLEAPFLDVLNTMMDTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPYQ 565
Query 117 NINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAV---------GYSSP 163
NI P+ P++ + ++D+RV + Y +L+ A+ GY +P
Sbjct 566 NIKPQ-----HYPSIHITAYENDERVPLKGIVSYTEKLKEAIAEHAKDTGEGYQTP 616
> tpv:TP04_0365 protease II (EC:3.4.21.83); K01354 oligopeptidase
B [EC:3.4.21.83]
Length=767
Score = 61.2 bits (147), Expect = 3e-09, Method: Composition-based stats.
Identities = 46/165 (27%), Positives = 78/165 (47%), Gaps = 24/165 (14%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGGE G W K K+ K S+ D ++L +K ++ +LAI S G+L G
Sbjct 566 HVRGGGELGEVWRKMGSKDKKYNSFYDLVDVIQFLISKNVTSRSKLAINVSSASGILGGC 625
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDKEEDFKYI----------Y 110
N RPDL + K+ +D A V+D P E +++ Y
Sbjct 626 VYNMRPDLCSVISFKLPYLDHF---------ATVNDPSQPLVELEYEEFGSTQEYECEEY 676
Query 111 KISPIHNINPEV----VRSDKQPAVLVMTADHDDRVSPFHSLKYI 151
+ I++++P V V S K+P++L+ +D R +H+ K++
Sbjct 677 NVDHIYSMDPCVNMKKVHS-KRPSILINCNINDVRAPWYHAAKFL 720
> ath:AT5G66960 prolyl oligopeptidase family protein
Length=792
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 52/158 (32%), Positives = 77/158 (48%), Gaps = 8/158 (5%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
++RGGG G W++ K S D+ A+YL ++LA G S GGL+V +
Sbjct 596 DVRGGGGKGKKWHQDGRGAKKLNSIKDYIQCAKYLVENNIVEENKLAGWGYSAGGLVVAS 655
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY---GNPDKEEDFKYIYKISPIHN 117
N PDLF+A V KV +D + I DY G P DF I + SP N
Sbjct 656 AINHCPDLFQAAVLKVPFLDPTHTLIYPILPLTAEDYEEFGYPGDINDFHAIREYSPYDN 715
Query 118 INPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQ 155
I +V+ PAVLV T+ + R + + K++A+++
Sbjct 716 IPKDVL----YPAVLV-TSSFNTRFGVWEAAKWVARVR 748
> bbo:BBOV_II002340 18.m06190; hypothetical protein; K01354 oligopeptidase
B [EC:3.4.21.83]
Length=487
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 38/155 (24%), Positives = 64/155 (41%), Gaps = 9/155 (5%)
Query 2 IRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGAC 61
+RGGGE G W+ A+K K D E+L + +A+ S G +L
Sbjct 293 VRGGGELGHIWHSGAVKSFKYKGVYDLIDVIEFLIANNMAQRSSIALSSTSAGAILAACV 352
Query 62 ANQRPDLFRAVVAKVGVMDL---LRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNI 118
N RPDL + K+ +D+ + + ++GN E + + +Y SP N
Sbjct 353 YNMRPDLCNCISLKLPFLDVFGSIHDTNEPLSSLEKEEFGN---ESNVECVYSYSPCDNT 409
Query 119 NPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQ 153
+P++++ D R H++ YI Q
Sbjct 410 GEP---GHLKPSLIIQCNSDDTRAPLRHTVSYIRQ 441
> dre:678599 MGC136971, si:dkey-16c7.3, wu:fc20g05; zgc:136971
Length=714
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 17/122 (13%)
Query 43 PDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---------VGVMDLLRFNKFTIGHAW 93
PD++A+MGGS+GG L Q PD +RA A+ +G D++ + ++G +
Sbjct 561 PDRVAVMGGSHGGFLACHLVGQYPDFYRACAARNPVINAATLLGTSDIVDWRYSSVGLQY 620
Query 94 VSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQ 153
D P + + K IH P++ + VL+M + D RVSP L+
Sbjct 621 AFDR-LPTSQSLISMLDKSPIIH--APQI-----RAPVLLMLGERDRRVSPHQGLELYRA 672
Query 154 LQ 155
L+
Sbjct 673 LK 674
> ath:AT4G14570 acylaminoacyl-peptidase-related; K01303 acylaminoacyl-peptidase
[EC:3.4.19.1]
Length=764
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/164 (23%), Positives = 63/164 (38%), Gaps = 14/164 (8%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N RG YG ++ + D A ++ G + P ++ ++GGS+GG L
Sbjct 567 NYRGSLGYGEDALQSLPGKVGSQDVKDCLLAVDHAIEMGIADPSRITVLGGSHGGFLTTH 626
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWV--------SDYGNPDKEEDFKYIYKI 112
Q PD F A A+ V ++ T W S Y ED +++
Sbjct 627 LIGQAPDKFVAAAARNPVCNMASMVGITDIPDWCFFEAYGDQSHYTEAPSAEDLSRFHQM 686
Query 113 SPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQH 156
SPI +I S + L + D RV + +Y+ L+
Sbjct 687 SPISHI------SKVKTPTLFLLGTKDLRVPISNGFQYVRALKE 724
> cel:F01F1.5 dpf-4; Dipeptidyl Peptidase Four (IV) family member
(dpf-4)
Length=629
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 0/88 (0%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N RG +G + + K DD + A+ L +G +++ I G S+GG L+ +
Sbjct 424 NYRGSTGFGTEFRRMLYKNCGVADRDDMLNGAKALVEQGKVDAEKVLITGSSSGGYLILS 483
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFT 88
C ++ +A V+ GV DLL ++ T
Sbjct 484 CLISPKNIIKAAVSVYGVADLLALDEDT 511
> ath:AT5G36210 serine-type peptidase
Length=730
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 56/206 (27%), Positives = 92/206 (44%), Gaps = 24/206 (11%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N G YG + + +++ V DD A+YL + G + +L I GGS GG A
Sbjct 528 NYGGSTGYGREYRERLLRQWGIVDVDDCCGCAKYLVSSGKADVKRLCISGGSAGGYTTLA 587
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNP--DKEEDFKYIYKISPIHNI 118
R D+F+A + GV DL + GH + S Y + E+DF Y+ SPI+ +
Sbjct 588 SLAFR-DVFKAGASLYGVADLKMLKE--EGHKFESRYIDNLVGDEKDF---YERSPINFV 641
Query 119 NPEVVRSDKQPAVLVMTADHDDR-VSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDTKAG 177
DK +++ +D+ V+P S K L+ PV+ L+ + + G
Sbjct 642 -------DKFSCPIILFQGLEDKVVTPDQSRKIYEALKKK---GLPVA--LVEYEGEQHG 689
Query 178 HGGGKPLMKVLDEQALTYGFIASVLG 203
+ + L++Q + F A V+G
Sbjct 690 FRKAENIKYTLEQQMV---FFARVVG 712
> mmu:235606 Apeh, MGC38101; acylpeptide hydrolase (EC:3.4.19.1);
K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=732
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 36/138 (26%), Positives = 58/138 (42%), Gaps = 17/138 (12%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A + + + + ++A+MGGS+GG L Q P+ + A +A+ V++++
Sbjct 562 DVQFAVQQVLQEEHFDARRVALMGGSHGGFLSCHLIGQYPETYSACIARNPVINIVSMMG 621
Query 87 FTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQP---------AVLVMTAD 137
T W E F Y P N+ E++ DK P VL+M
Sbjct 622 TTDIPDWCM------VETGFPYSNDYLPDLNVLEEML--DKSPIKYIPQVKTPVLLMLGQ 673
Query 138 HDDRVSPFHSLKYIAQLQ 155
D RV L+Y L+
Sbjct 674 EDRRVPFKQGLEYYHALK 691
> hsa:327 APEH, ACPH, APH, D3F15S2, D3S48E, DNF15S2, MGC2178,
OPH; N-acylaminoacyl-peptide hydrolase (EC:3.4.19.1); K01303
acylaminoacyl-peptidase [EC:3.4.19.1]
Length=732
Score = 39.3 bits (90), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 38/126 (30%), Positives = 59/126 (46%), Gaps = 14/126 (11%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A E + + + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 87 FTIGHAW-VSDYGNP---DKEEDFKYIYKI---SPIHNINPEVVRSDKQPAVLVMTADHD 139
T W V + G P D D ++ SPI I P+V K P +L++ +
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYI-PQV----KTPLLLML--GQE 674
Query 140 DRVSPF 145
DR PF
Sbjct 675 DRRVPF 680
> ath:AT5G24260 prolyl oligopeptidase family protein; K01278 dipeptidyl-peptidase
4 [EC:3.4.14.5]
Length=746
Score = 38.5 bits (88), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 40/157 (25%), Positives = 71/157 (45%), Gaps = 15/157 (9%)
Query 3 RGGGEYGVGWYKAAIKENK-QVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGAC 61
RG G+ +++ +K N V +D + A++L +G + PD + + G S GG L
Sbjct 566 RGTARRGLK-FESWMKHNCGYVDAEDQVTGAKWLIEQGLAKPDHIGVYGWSYGGYLSATL 624
Query 62 ANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYKISPIHNINP 120
+ P++F V+ V ++ F + Y G P +EE + K S +H++
Sbjct 625 LTRYPEIFNCAVSGAPVTSWDGYDSF-----YTEKYMGLPTEEERY---LKSSVMHHVGN 676
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA 157
+DKQ +LV D+ V H+ + + L A
Sbjct 677 ---LTDKQKLMLVHGMI-DENVHFRHTARLVNALVEA 709
> mmu:269109 Dpp10, 6430601K09Rik, DPP_X, Dprp3; dipeptidylpeptidase
10
Length=800
Score = 38.1 bits (87), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 50/181 (27%), Positives = 73/181 (40%), Gaps = 20/181 (11%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACA 62
RG G G+ + + V D +A +YL + Y +L+I G GG +
Sbjct 606 RGSGFQGLKVLQEIHRRIGSVEAKDQVAAVKYLLKQPYIDSKRLSIFGKGYGGYIASMIL 665
Query 63 NQRPDLFR--AVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYKISPI-HNI 118
F+ AVVA + M L A+ Y G P KEE Y+ S + HNI
Sbjct 666 KSDEKFFKCGAVVAPISDMKLY-------ASAFSERYLGMPSKEES---TYQASSVLHNI 715
Query 119 NPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA-VGYSSPVSTPLIAHIDTKAG 177
+ K+ +L++ D +V HS + I L A V Y+ V HI K+
Sbjct 716 H-----GLKEENLLIIHGTADTKVHFQHSAELIKHLIKAGVNYTLQVYPDEGYHISDKSK 770
Query 178 H 178
H
Sbjct 771 H 771
> dre:321028 apeh, cb5, sb:cb5, wu:fi37d02; acylpeptide hydrolase
(EC:3.4.19.1); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=730
Score = 37.4 bits (85), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 57/136 (41%), Gaps = 13/136 (9%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A + + +G ++A++GGS+GG L Q P ++A VA+ V +L
Sbjct 560 DVQFAVDSVLKQGGFDEQKVAVIGGSHGGFLACHLIGQYPGFYKACVARNPVTNLASMVC 619
Query 87 FTIGHAWVSDYGNPDKEEDFKYIYKI-------SPIHNINPEVVRSDKQPAVLVMTADHD 139
T W D + D + I SPI ++ + + VL+M + D
Sbjct 620 CTDIPDWCIVEAGFDYKPDIQLEPAILEQMLIKSPIKHV------AKVKTPVLLMLGEGD 673
Query 140 DRVSPFHSLKYIAQLQ 155
RV ++Y L+
Sbjct 674 KRVPNKQGIEYYKALK 689
> xla:100190777 dpp8-a, dp8, dpp8, dpp8a, dprp1; dipeptidyl-peptidase
8; K08655 dipeptidyl-peptidase 8 [EC:3.4.14.5]
Length=888
Score = 37.4 bits (85), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 31/109 (28%), Positives = 48/109 (44%), Gaps = 7/109 (6%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTK-GYSTPDQLAIMGGSNGGLLVGAC 61
RG G+ + A + QV DD +YL K + D++ + G S GG L
Sbjct 690 RGSCHRGLKFEGAFKYKMGQVEIDDQVEGLQYLAAKHSFIDLDRVGVHGWSYGGYLSLMA 749
Query 62 ANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYI 109
QRPD+FR +A V + ++ + Y G+PD+ E Y+
Sbjct 750 LVQRPDIFRVAIAGAPVTLWIFYDT-----GYTERYMGHPDQNEHGYYL 793
> dre:100333019 N-acylaminoacyl-peptide hydrolase-like
Length=742
Score = 37.0 bits (84), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 31/135 (22%), Positives = 60/135 (44%), Gaps = 9/135 (6%)
Query 27 DFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFNK 86
D Q A + + +G ++A++GGS+GG L Q P ++A VA+ V++L
Sbjct 572 DVQFAVDSVLKQGGFDEQKVAVIGGSHGGFLACHLIGQYPGFYKACVARNPVINLTSM-- 629
Query 87 FTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPA-----VLVMTADHDDR 141
+G + D+ + +K + P + + + K A VL++ + D R
Sbjct 630 --VGGTDIPDWCTVEAGYKYKPDVYLEPAVLVQMLIKKKIKHVAKVKTPVLLLLGEDDKR 687
Query 142 VSPFHSLKYIAQLQH 156
V ++Y L++
Sbjct 688 VPNKQGIEYYRALKN 702
> cel:R11E3.8 dpf-5; Dipeptidyl Peptidase Four (IV) family member
(dpf-5); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=737
Score = 36.6 bits (83), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 36/164 (21%), Positives = 66/164 (40%), Gaps = 16/164 (9%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKG-YSTPDQLAIMGGSNGGLLVG 59
N RG +G + +A + D +A + K + D++ + GGS+GG LV
Sbjct 540 NFRGSLGFGDDFIRALPGNCGDMDVKDVHNAVLTVLDKNPRISRDKVVLFGGSHGGFLVS 599
Query 60 ACANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWV--------SDYGNPDKEEDFKYIYK 111
Q P +++ VA V+++ + T W D+ E + ++
Sbjct 600 HLIGQYPGFYKSCVALNPVVNIATMHDITDIPEWCYFEGTGEYPDWTKITTTEQREKMFN 659
Query 112 ISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQ 155
SPI ++ + L++ + D RV P H +I L+
Sbjct 660 SSPIAHV------ENATTPYLLLIGEKDLRVVP-HYRAFIRALK 696
> tgo:TGME49_046800 acylamino-acid-releasing enzyme, putative
(EC:1.7.2.2 3.4.19.1); K01303 acylaminoacyl-peptidase [EC:3.4.19.1]
Length=851
Score = 35.0 bits (79), Expect = 0.21, Method: Composition-based stats.
Identities = 33/125 (26%), Positives = 49/125 (39%), Gaps = 10/125 (8%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYS--TPDQLAIMGGSNGGLLV 58
N RG +G + + + + DD + A L TP + ++GGS+GG L
Sbjct 638 NYRGSLGFGQEELLSLLGKAGRQDVDDVKEAVSDLIASDPDAYTPARTVVVGGSHGGFLT 697
Query 59 GACANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDK--------EEDFKYIY 110
Q PDLF A + V +L + W + G K E D +Y
Sbjct 698 CHLIGQFPDLFAAASTRNPVTNLASMVVESDIPDWCAAEGLHRKFHPSFGLTENDIVALY 757
Query 111 KISPI 115
K SP+
Sbjct 758 KASPV 762
> cel:F44B9.1 dpf-6; Dipeptidyl Peptidase Four (IV) family member
(dpf-6)
Length=740
Score = 34.7 bits (78), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 48/211 (22%), Positives = 81/211 (38%), Gaps = 23/211 (10%)
Query 1 NIRGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGA 60
N RG +G A E + + D A E+ +KG + ++A+MGGS GG
Sbjct 465 NFRGSTGFGKRLTNAGNGEWGRKMHFDILDAVEFAVSKGIANRSEVAVMGGSYGGYETLV 524
Query 61 CANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDYGNPDK--------EEDFKYIYKI 112
P F V VG +L+ + I W+ + K EE + +
Sbjct 525 ALTFTPQTFACGVDIVGPSNLISLVQ-AIPPYWLGFRKDLIKMVGADISDEEGRQSLQSR 583
Query 113 SPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHI 172
SP+ + R K ++++ +D RV S +++A L+ P+ +
Sbjct 584 SPLFFAD----RVTK--PIMIIQGANDPRVKQAESDQFVAALEKK-------HIPVTYLL 630
Query 173 DTKAGHGGGKPLMKVLDEQALTYGFIASVLG 203
GHG KP +++ F+ LG
Sbjct 631 YPDEGHGVRKP-QNSMEQHGHIETFLQQCLG 660
> mmu:74388 Dpp8, 2310004I03Rik, 4932434F09Rik, AI666706, DPP_VIII;
dipeptidylpeptidase 8 (EC:3.4.14.5); K08655 dipeptidyl-peptidase
8 [EC:3.4.14.5]
Length=892
Score = 33.5 bits (75), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 48/109 (44%), Gaps = 7/109 (6%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTK-GYSTPDQLAIMGGSNGGLLVGAC 61
RG G+ + A + Q+ DD +YL ++ + D++ I G S GG L
Sbjct 699 RGSCHRGLKFEGAFKYKMGQIEIDDQVEGLQYLASQYDFIDLDRVGIHGWSYGGYLSLMA 758
Query 62 ANQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYI 109
QR D+FR +A V + ++ + Y G+PD+ E Y+
Sbjct 759 LMQRSDIFRVAIAGAPVTLWIFYDT-----GYTERYMGHPDQNEQGYYL 802
> xla:432175 dpp10, MGC84485; dipeptidyl-peptidase 10 (non-functional)
Length=796
Score = 33.1 bits (74), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 69/165 (41%), Gaps = 16/165 (9%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACA 62
RG G G+ + + V D +A E+L + + P++L+I G GG +
Sbjct 601 RGSGFQGLKILQEVHRGLGSVEVKDQIAAVEWLLKEPFIDPNRLSIFGKGYGGYIASMIL 660
Query 63 NQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYKISPI-HNINP 120
LF+ + D+ + A+ Y G P +EE Y+ S + HN++
Sbjct 661 KANDRLFKCGALFAPITDMRLY-----ASAFSERYLGLPSREE---ITYQASSVLHNVH- 711
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA-VGYSSPV 164
V DK +L++ D +V HS + I L A V Y+ V
Sbjct 712 --VLKDKN--LLLIHGTADAKVHFQHSAELIKHLVKAGVNYTMQV 752
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 40/162 (24%), Positives = 65/162 (40%), Gaps = 17/162 (10%)
Query 20 NKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---V 76
++Q D Q+A ++L + ++ + G S GG + PD A++ +
Sbjct 127 SQQGIIKDAQAALDHLSGRTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALILENTFT 186
Query 77 GVMD----LLRFNKFTIGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVL 132
++D LL F K+ IG G+ K SP I + + KQP VL
Sbjct 187 SILDMAGVLLPFLKWFIG-------GSGTKSLKLLNFVVRSPWKTI--DAIAEIKQP-VL 236
Query 133 VMTADHDDRVSPFHSLKYIAQLQHAVGYSSPVSTPLIAHIDT 174
++ D+ V PFH A+ + V P H+DT
Sbjct 237 FLSGLQDEMVPPFHMKMLYAKAAARNPQCTFVEFPSGMHMDT 278
> hsa:57628 DPP10, DPL2, DPPY, DPRP3; dipeptidyl-peptidase 10
(non-functional)
Length=789
Score = 32.0 bits (71), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 64/165 (38%), Gaps = 16/165 (9%)
Query 3 RGGGEYGVGWYKAAIKENKQVSYDDFQSAAEYLFTKGYSTPDQLAIMGGSNGGLLVGACA 62
RG G G+ + + V D +A ++L Y +L+I G GG +
Sbjct 595 RGSGFQGLKILQEIHRRLGSVEVKDQITAVKFLLKLPYIDSKRLSIFGKGYGGYIASMIL 654
Query 63 NQRPDLFRAVVAKVGVMDLLRFNKFTIGHAWVSDY-GNPDKEEDFKYIYK-ISPIHNINP 120
LF+ + DL + A+ Y G P KEE Y+ S +HN
Sbjct 655 KSDEKLFKCGSVVAPITDLKLY-----ASAFSERYLGMPSKEES---TYQAASVLHN--- 703
Query 121 EVVRSDKQPAVLVMTADHDDRVSPFHSLKYIAQLQHA-VGYSSPV 164
V K+ +L++ D +V HS + I L A V Y+ V
Sbjct 704 --VHGLKEENILIIHGTADTKVHFQHSAELIKHLIKAGVNYTMQV 746
> hsa:64787 EPS8L2, EPS8R2, FLJ16738, FLJ21935, FLJ22171, MGC126530,
MGC3088; EPS8-like 2
Length=715
Score = 30.0 bits (66), Expect = 5.9, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 34/80 (42%), Gaps = 1/80 (1%)
Query 89 IGHAWVSDYGNPDKEEDFKYIYKISPIHNINPEVVRSDKQPAVLVMTADHDDRVSPFHSL 148
+G A D G P ++ KY SP H + P +K + M +D+ + ++
Sbjct 546 LGEARPEDAGAPFEQAGQKYWGPASPTHKLPPS-FPGNKDELMQHMDEVNDELIRKISNI 604
Query 149 KYIAQLQHAVGYSSPVSTPL 168
+ Q V S PVS PL
Sbjct 605 RAQPQRHFRVERSQPVSQPL 624
Lambda K H
0.317 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6880722740
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40