bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1213_orf2
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
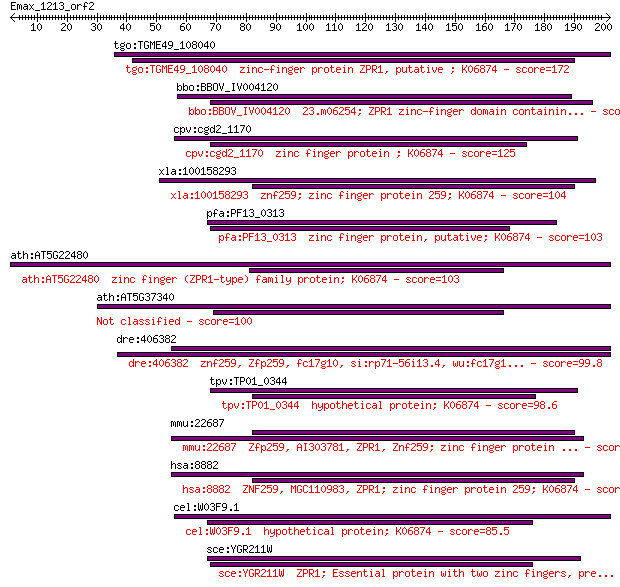
tgo:TGME49_108040 zinc-finger protein ZPR1, putative ; K06874 172 9e-43
bbo:BBOV_IV004120 23.m06254; ZPR1 zinc-finger domain containin... 126 4e-29
cpv:cgd2_1170 zinc finger protein ; K06874 125 1e-28
xla:100158293 znf259; zinc finger protein 259; K06874 104 2e-22
pfa:PF13_0313 zinc finger protein, putative; K06874 103 3e-22
ath:AT5G22480 zinc finger (ZPR1-type) family protein; K06874 103 4e-22
ath:AT5G37340 zinc finger (ZPR1-type) family protein 100 2e-21
dre:406382 znf259, Zfp259, fc17g10, si:rp71-56i13.4, wu:fc17g1... 99.8 6e-21
tpv:TP01_0344 hypothetical protein; K06874 98.6 1e-20
mmu:22687 Zfp259, AI303781, ZPR1, Znf259; zinc finger protein ... 93.6 4e-19
hsa:8882 ZNF259, MGC110983, ZPR1; zinc finger protein 259; K06874 92.8 7e-19
cel:W03F9.1 hypothetical protein; K06874 85.5 1e-16
sce:YGR211W ZPR1; Essential protein with two zinc fingers, pre... 84.0 3e-16
> tgo:TGME49_108040 zinc-finger protein ZPR1, putative ; K06874
Length=568
Score = 172 bits (435), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 86/166 (51%), Positives = 106/166 (63%), Gaps = 8/166 (4%)
Query 36 AGAAAGAAGGEEEDCSSSLLREEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSC 95
A A G G E+ED SL PV PHCGT+ N VC++ +PGFR CL+F+F C
Sbjct 292 ASPAEGREGEEKEDYLFSL--------PVSCPHCGTEGSNNVCEIDVPGFRRCLIFSFLC 343
Query 96 SICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQA 155
CG ++S IK AGA+G GR+WIL VE EDLNRDVLK DT+ + IPSLDFSM GG Q
Sbjct 344 QSCGGRHSEIKAAGAFGAVGRKWILNVETAEDLNRDVLKSDTAVVEIPSLDFSMRGGVQG 403
Query 156 GTLTTIEGLIKSLREGLAESVPFAVGDSAAAASRERMGCIGTYLQR 201
G TT+EGL+ L L +S PFA GDSA RE++ + LQ+
Sbjct 404 GEFTTVEGLLGKLATALGDSAPFACGDSAPQEKREKLSELIGKLQK 449
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/148 (26%), Positives = 70/148 (47%), Gaps = 5/148 (3%)
Query 42 AAGGEEEDCSSSLLREEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAK 101
A EE+D +L E+L P+C + +P F+E +L +FSC C
Sbjct 25 ARTAEEDDLLQNLTEVESLC-----PNCEENGTTLLLLHKVPHFKEIVLISFSCPHCHYS 79
Query 102 NSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTI 161
N +++A G R LTV+ DL+R +++ + +T+I+ ++ + G LTT+
Sbjct 80 NREVQSAACLAPQGVRLELTVQSAADLDRQIVRSEHATLIVKEVELEVPPKRDRGELTTV 139
Query 162 EGLIKSLREGLAESVPFAVGDSAAAASR 189
EG I+ + + L P ++ A +
Sbjct 140 EGAIRRMIDALRAGQPVRRAEAPEVAEK 167
> bbo:BBOV_IV004120 23.m06254; ZPR1 zinc-finger domain containing
protein; K06874
Length=453
Score = 126 bits (317), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 56/132 (42%), Positives = 83/132 (62%), Gaps = 0/132 (0%)
Query 57 EEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGR 116
+E L LPV+ PHCG +NK+C++ +PGF C++ AF+C CGAK++ IK G Y + R
Sbjct 249 KEGLSLPVDCPHCGKVGNNKICEVLVPGFGPCVIMAFTCENCGAKSNEIKPGGGYKEHAR 308
Query 117 RWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESV 176
+W L V+ DLNRDV+ +T+TI IP L+ M G TT+EG++ + L +
Sbjct 309 KWTLKVQDVTDLNRDVIISETATIHIPLLELDMTAGTIGAVYTTVEGMLIKHADSLETAY 368
Query 177 PFAVGDSAAAAS 188
PF +GDSA ++
Sbjct 369 PFLLGDSADPSN 380
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 58/128 (45%), Gaps = 5/128 (3%)
Query 68 HCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEED 127
CG V IP F+E +L +F C CG +NS ++ A G R V E
Sbjct 30 QCGAMGITMVLMHMIPHFKEVILMSFECPSCGYRNSELQDAAPLQDYGLRLKAHVAYEGA 89
Query 128 LNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAA 187
L V+ T++ I +DF + + GT+TTIEG + L GL + + S A A
Sbjct 90 LRNQVVLSGTTSCRIEEIDFEFQPTMEKGTVTTIEGYLMRLAGGLGDHI-----SSIADA 144
Query 188 SRERMGCI 195
RE G I
Sbjct 145 MRENPGII 152
> cpv:cgd2_1170 zinc finger protein ; K06874
Length=475
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 57/135 (42%), Positives = 85/135 (62%), Gaps = 1/135 (0%)
Query 56 REEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTG 115
E+ + V P+CG + VC++ IPGFR CL+ AF C+ CG K + +K +GAYG+
Sbjct 264 NEDRIKFDVPCPNCGNNGESDVCEIDIPGFRRCLIMAFVCNFCGIKTNELKPSGAYGELA 323
Query 116 RRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAES 175
++WILTVE E DLNRD+LK DT++I IP ++ M G+ TT+EG+I + + L +
Sbjct 324 KKWILTVESELDLNRDILKSDTASIEIPEIELEMGMGSLGSLFTTVEGMIVKITDSLKDC 383
Query 176 VPFAVGDSAAAASRE 190
F GDSA + ++
Sbjct 384 FTFQ-GDSATSEQKK 397
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 55/106 (51%), Gaps = 0/106 (0%)
Query 68 HCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEED 127
+C + K+ IP FR+ +L +F C CG KN+ I++ G G L V D
Sbjct 24 NCHKEGETKLLLTSIPQFRDVILMSFECPHCGFKNNEIQSGGVLQDKGECIELVVTNVSD 83
Query 128 LNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLA 173
L+R ++K + +TI I + + Q G ++TIEG+I +GL+
Sbjct 84 LDRQIVKSEFATISILEQELDIPPSTQKGVISTIEGIITKTIQGLS 129
> xla:100158293 znf259; zinc finger protein 259; K06874
Length=446
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 55/148 (37%), Positives = 85/148 (57%), Gaps = 4/148 (2%)
Query 51 SSSLLREEALLLPVERPHCGT--QTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTA 108
+S LR+E L P P C +T+ K+ Q IP F+E ++ A +C CG + + +K+
Sbjct 234 TSEELRDEVLQFPTNCPECNVPAETNMKLVQ--IPHFKEVVIMATNCDSCGHRTNEVKSG 291
Query 109 GAYGQTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSL 168
GA G + L + DL RDVLK +T +I IP L+F + GA G TT+EGL+K +
Sbjct 292 GAIEPLGTKITLHITDLSDLTRDVLKSETCSIGIPELEFELGMGALGGKFTTLEGLLKDI 351
Query 169 REGLAESVPFAVGDSAAAASRERMGCIG 196
++ + + PF VGDS+ + RE++ G
Sbjct 352 KDLVVDKNPFTVGDSSTSDRREKLEEFG 379
Score = 86.3 bits (212), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 66/108 (61%), Gaps = 0/108 (0%)
Query 82 IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTII 141
+P F+E ++ +F+C CG N+ I++AG + G R+ L+V ++D+NR+V+K D +T
Sbjct 51 VPFFKEIIVSSFTCDSCGWSNTEIQSAGRIQEQGVRYSLSVRSKQDVNREVIKTDYATTQ 110
Query 142 IPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAASR 189
IP LDF + Q G LTTIEG+++ GL + P ++ + A +
Sbjct 111 IPELDFEIPACTQKGALTTIEGILERTIAGLQQEQPLRRAENESVADK 158
> pfa:PF13_0313 zinc finger protein, putative; K06874
Length=613
Score = 103 bits (257), Expect = 3e-22, Method: Composition-based stats.
Identities = 49/117 (41%), Positives = 71/117 (60%), Gaps = 0/117 (0%)
Query 67 PHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEE 126
P C N C++ IPGF++CL+ +F C C K S IK++G G++ LTV +
Sbjct 402 PCCNHMGMNNFCEINIPGFKKCLILSFVCPNCNFKTSEIKSSGEINPKGKKITLTVNNKN 461
Query 127 DLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDS 183
DLNR V+K +T++I IP ++ + + G GTLTT+EGLI + E L E F +GDS
Sbjct 462 DLNRFVIKSETASINIPVVELTSDYGTLGGTLTTVEGLIMKIIESLEEKFKFLLGDS 518
Score = 65.5 bits (158), Expect = 1e-10, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 54/100 (54%), Gaps = 0/100 (0%)
Query 68 HCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEED 127
+C + NK+ ++ IP F+ L+ +F C C KN+VI+ G + + + +E
Sbjct 26 NCEKEGLNKIVKINIPYFKNVLIHSFECEFCNYKNNVIQDLNQIKDKGVKISMKINNKEL 85
Query 128 LNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKS 167
L+R ++K + + IP +DF + Q G++ TIEG + +
Sbjct 86 LDRQLIKSEYGVLKIPEIDFEIPKETQKGSINTIEGFLHT 125
> ath:AT5G22480 zinc finger (ZPR1-type) family protein; K06874
Length=493
Score = 103 bits (257), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 72/208 (34%), Positives = 102/208 (49%), Gaps = 12/208 (5%)
Query 1 GESASSGESGESASGDTQAAGAAGAAGAAGAAGAAAGAAAGAAGGEEEDCSSSLLR---- 56
G A+ ++G+S G A A G GA AG A A D S +L R
Sbjct 219 GYVANPSQAGQS-EGSLGAPSTKTAYVPNGTIGATAGHRA-IAQSNSTDISDNLFRYSAP 276
Query 57 EEALLLPVERPHCGTQTHNKVCQLF---IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQ 113
EE + P CG T ++F IP F+E ++ A +C CG +NS +K GA +
Sbjct 277 EEVMTFPST---CGACTEPCETRMFVTKIPYFQEVIVMASTCDSCGYRNSELKPGGAIPE 333
Query 114 TGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLA 173
G++ L+V DL+RDV+K DT+ +IIP LD + GG G +TT+EGL+ +RE LA
Sbjct 334 KGKKITLSVRNITDLSRDVIKSDTAGVIIPELDLELAGGTLGGMVTTVEGLVTQIRESLA 393
Query 174 ESVPFAVGDSAAAASRERMGCIGTYLQR 201
F GDS + + G L +
Sbjct 394 RVHGFTFGDSMEESKLNKWREFGARLTK 421
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 52/87 (59%), Gaps = 2/87 (2%)
Query 81 FIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEG--EEDLNRDVLKGDTS 138
IP FR+ L+ AF C CG +N+ ++ AG G + L V + +R V+K +++
Sbjct 50 LIPHFRKVLISAFECPHCGERNNEVQFAGEIQPRGCCYNLEVLAGDVKIFDRQVVKSESA 109
Query 139 TIIIPSLDFSMEGGAQAGTLTTIEGLI 165
TI IP LDF + AQ G+L+T+EG++
Sbjct 110 TIKIPELDFEIPPEAQRGSLSTVEGIL 136
> ath:AT5G37340 zinc finger (ZPR1-type) family protein
Length=493
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 66/181 (36%), Positives = 95/181 (52%), Gaps = 15/181 (8%)
Query 30 GAAGAAAGAAAGAAGGEEEDCSSSLLR----EEALLLPVERPHCGTQTHNKVCQ--LF-- 81
G GA AG A A D S +L R EE + P CG T K+C+ +F
Sbjct 247 GTIGATAGHRA-IAQSNSTDISDNLFRYSAPEEVMTFPST---CGACT--KLCETRMFVT 300
Query 82 -IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTI 140
IP F+E ++ A +C CG +NS +K GA + G++ L+V+ DL+RDV+K DT+ +
Sbjct 301 KIPYFQEVIVMASTCDDCGYRNSELKPGGAIPEKGKKITLSVKNITDLSRDVIKSDTAGV 360
Query 141 IIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAASRERMGCIGTYLQ 200
IP LD + GG G +TT+EGL+ +RE LA F GDS + + G+ L
Sbjct 361 KIPELDLELAGGTLGGMVTTVEGLVTQIRESLARVHGFTFGDSLEQSKINKWKEFGSRLT 420
Query 201 R 201
+
Sbjct 421 K 421
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 56/99 (56%), Gaps = 2/99 (2%)
Query 69 CGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEG--EE 126
CG + IP FR+ L+ AF C CG +N+ ++ AG G + L V +
Sbjct 38 CGENGTTRFLLTLIPHFRKVLISAFECPHCGERNNEVQFAGEIQPRGCSYHLEVSAGDVK 97
Query 127 DLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLI 165
+R V+K +++TI IP LDF + AQ+G+L+T+EG++
Sbjct 98 TFDRQVVKSESATIKIPELDFEIPPEAQSGSLSTVEGIL 136
> dre:406382 znf259, Zfp259, fc17g10, si:rp71-56i13.4, wu:fc17g10,
zgc:56478; zinc finger protein 259; K06874
Length=441
Score = 99.8 bits (247), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 47/147 (31%), Positives = 81/147 (55%), Gaps = 0/147 (0%)
Query 55 LREEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQT 114
LR E L+ P C + + + IP F+E ++ A +C CG + + +K+ GA +
Sbjct 234 LRNEVLVFNTNCPECNAPANTNMKLVQIPHFKEVIIMATNCDSCGHRTNEVKSGGATEEL 293
Query 115 GRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAE 174
G R L + D++RD+LK +T +++IP L+F + A G TT+EGL+K ++E +
Sbjct 294 GTRITLHLTDPSDMSRDLLKSETCSVLIPELEFELGMAALGGKFTTVEGLLKDIKELIVS 353
Query 175 SVPFAVGDSAAAASRERMGCIGTYLQR 201
PF GDS+A+ E++ G + +
Sbjct 354 KNPFMCGDSSASDRVEKLQLFGQKIDK 380
Score = 82.4 bits (202), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 84/165 (50%), Gaps = 12/165 (7%)
Query 37 GAAAGAAGGEEEDCSSSLLREEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCS 96
G+ E+ED +++ E+L + +C ++ IP F+E ++ +F+C
Sbjct 11 GSVFKDINAEDEDQQPTVI--ESLCM-----NCYENGSTRLLLTKIPFFKEIIISSFACP 63
Query 97 ICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAG 156
C N+ I++AG + G ++ L V+ + D+NR+V+K D+++ IP LDF + Q G
Sbjct 64 HCNWSNTEIQSAGRIQEQGVQYTLQVKTKRDMNREVVKSDSASTRIPQLDFEIPAFTQKG 123
Query 157 TLTTIEGLIKSLREGLAESVPFAVGDSAAAASRERMGCIGTYLQR 201
+L+TIEGL+ GL + P A + I ++QR
Sbjct 124 SLSTIEGLLDRAVAGLEQDQPIRKATDPTVADK-----IEEFIQR 163
> tpv:TP01_0344 hypothetical protein; K06874
Length=595
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 69/123 (56%), Gaps = 0/123 (0%)
Query 68 HCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEED 127
+CG + N++CQ+ IPGF +C++ +F C C K + +K G + G+ W L + +D
Sbjct 332 NCGYKGKNQICQIVIPGFDKCIIMSFVCDNCDYKTNELKPGGGVKKYGKVWHLKINSIDD 391
Query 128 LNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAA 187
+ RD++ +T I I L+ + G+ + TTIEGLI + E L + PF +GDS+
Sbjct 392 IKRDIILSNTCDITINELELFISAGSLSSLFTTIEGLINKIIENLQSTFPFLIGDSSPYN 451
Query 188 SRE 190
E
Sbjct 452 KDE 454
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/119 (23%), Positives = 50/119 (42%), Gaps = 24/119 (20%)
Query 82 IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTII 141
IP F + L+ +F CS C KN+ I G + + V E LN ++ +TS +
Sbjct 67 IPHFNDILVMSFECSFCNNKNNEILNIAKLQNHGVSYNIHVNNLEGLNNQIVITNTSAVK 126
Query 142 IPS------------------------LDFSMEGGAQAGTLTTIEGLIKSLREGLAESV 176
+ + L+F + + G +TTIEGL+ ++ L++ +
Sbjct 127 LSNENFHIILVVISYITSIYSKFIQFELEFEIPKLDRKGIVTTIEGLLTNIINNLSDHI 185
> mmu:22687 Zfp259, AI303781, ZPR1, Znf259; zinc finger protein
259; K06874
Length=459
Score = 93.6 bits (231), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/108 (43%), Positives = 64/108 (59%), Gaps = 0/108 (0%)
Query 82 IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTII 141
IP FRE ++ +FSC CG N+ I++AG G R+ LTV +ED+NR+V+K D++T
Sbjct 67 IPFFREIIVSSFSCEHCGWNNTEIQSAGRIQDQGVRYTLTVRSQEDMNREVVKTDSATTR 126
Query 142 IPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAASR 189
IP LDF + +Q G LTT+EGLI GL + P A A R
Sbjct 127 IPELDFEIPAFSQKGALTTVEGLISRAISGLEQDQPTRRAVEGAIAER 174
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 75/140 (53%), Gaps = 5/140 (3%)
Query 55 LREEALLLPVERPHCGT--QTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYG 112
LR E L P C QT+ K+ Q IP F+E ++ A +C CG + + +K+ GA
Sbjct 248 LRNEVLQFNTNCPECNAPAQTNMKLVQ--IPHFKEVIIMATNCENCGHRTNEVKSGGAVE 305
Query 113 QTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGL 172
G R L + D+ RD+LK +T ++ IP L+F + G TT+EGL+K +RE L
Sbjct 306 PLGTRITLHITDPSDMTRDLLKSETCSVEIPELEFELGMAVLGGKFTTLEGLLKDIRE-L 364
Query 173 AESVPFAVGDSAAAASRERM 192
PF +GDS+ E++
Sbjct 365 VTKNPFTLGDSSNPDQSEKL 384
> hsa:8882 ZNF259, MGC110983, ZPR1; zinc finger protein 259; K06874
Length=459
Score = 92.8 bits (229), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 51/140 (36%), Positives = 75/140 (53%), Gaps = 5/140 (3%)
Query 55 LREEALLLPVERPHCGT--QTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYG 112
LR E L P C QT+ K+ Q IP F+E ++ A +C CG + + +K+ GA
Sbjct 248 LRNEVLQFSTNCPECNAPAQTNMKLVQ--IPHFKEVIIMATNCENCGHRTNEVKSGGAVE 305
Query 113 QTGRRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGL 172
G R L + D+ RD+LK +T ++ IP L+F + G TT+EGL+K +RE L
Sbjct 306 PLGTRITLHITDASDMTRDLLKSETCSVEIPELEFELGMAVLGGKFTTLEGLLKDIRE-L 364
Query 173 AESVPFAVGDSAAAASRERM 192
PF +GDS+ ER+
Sbjct 365 VTKNPFTLGDSSNPGQTERL 384
Score = 90.1 bits (222), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 45/108 (41%), Positives = 63/108 (58%), Gaps = 0/108 (0%)
Query 82 IPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEEDLNRDVLKGDTSTII 141
IP FRE ++ +FSC CG N+ I++AG G R+ L+V ED+NR+V+K D++
Sbjct 67 IPFFREIIVSSFSCEHCGWNNTEIQSAGRIQDQGVRYTLSVRALEDMNREVVKTDSAATR 126
Query 142 IPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAAASR 189
IP LDF + +Q G LTT+EGLI GL + P + A A R
Sbjct 127 IPELDFEIPAFSQKGALTTVEGLITRAISGLEQDQPARRANKDATAER 174
> cel:W03F9.1 hypothetical protein; K06874
Length=455
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/146 (33%), Positives = 78/146 (53%), Gaps = 3/146 (2%)
Query 56 REEALLLPVERPHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTG 115
++E L + P+C T K+ IP F+ ++ + +C CG K++ +K+ GA G
Sbjct 237 KQEVLHFATDCPNCHGPTEVKMKPTDIPFFQTVIIMSLACDRCGYKSNEVKSGGAIRDQG 296
Query 116 RRWILTVEGEEDLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAES 175
R + +E + DL RDVLK DT + IP +D + G A G TTIEGL+ + +E L
Sbjct 297 CRMSVKLEKDLDLARDVLKTDTCALSIPEIDLEVGGNALCGRFTTIEGLLTATKEQLDAQ 356
Query 176 VPFAVGDSAAAASRERMGCIGTYLQR 201
F +GDSA + + T+L++
Sbjct 357 SSFFMGDSAQTGEK---SAVTTFLEK 379
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 60/109 (55%), Gaps = 0/109 (0%)
Query 67 PHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEE 126
P C ++ IP +R +L +F C CG KN+ I++ A + G +L V+ E
Sbjct 29 PVCEEDGETRIMCTSIPYYRAVILMSFECPHCGHKNNEIQSGEAVQEHGTLIVLRVQKPE 88
Query 127 DLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAES 175
DL R ++K + ++I +P L + +Q G +TT+EG+++ + GL++
Sbjct 89 DLRRQLVKSEYASIEVPELQLEIPHKSQPGEVTTVEGVLERVHRGLSQD 137
> sce:YGR211W ZPR1; Essential protein with two zinc fingers, present
in the nucleus of growing cells but relocates to the
cytoplasm in starved cells via a process mediated by Cpr1p;
binds to translation elongation factor eEF-1 (Tef1p); K06874
Length=486
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/125 (32%), Positives = 67/125 (53%), Gaps = 0/125 (0%)
Query 67 PHCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEE 126
P C + + + IP F+E ++ + C CG K++ +KT GA GRR L +
Sbjct 296 PSCTQECETHMKPVNIPHFKEVIIMSTVCDHCGYKSNEVKTGGAIPDKGRRITLYCDDAA 355
Query 127 DLNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAESVPFAVGDSAAA 186
DL+RD+LK +T +++IP L ++ G G TT+EGL++ + E L + DS
Sbjct 356 DLSRDILKSETCSMVIPELHLDIQEGTLGGRFTTLEGLLRQVYEELESRIFTQTSDSMDE 415
Query 187 ASRER 191
A++ R
Sbjct 416 ATKAR 420
Score = 78.6 bits (192), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 61/108 (56%), Gaps = 2/108 (1%)
Query 68 HCGTQTHNKVCQLFIPGFRECLLFAFSCSICGAKNSVIKTAGAYGQTGRRWILTVEGEED 127
+CG ++ IP FRE ++ +F C CG KN I+ A + G R++L VE ED
Sbjct 56 NCGKNGTTRLLLTSIPYFREIIIMSFDCPHCGFKNCEIQPASQIQEKGSRYVLKVECRED 115
Query 128 LNRDVLKGDTSTIIIPSLDFSMEGGAQAGTLTTIEGLIKSLREGLAES 175
NR V+K +T+T LD +E A+ G LTT+EGL+ + + L++
Sbjct 116 FNRQVIKSETATCKFVELD--IEIPAKRGQLTTVEGLLSEMIDDLSQD 161
Lambda K H
0.313 0.130 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6060209704
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40