bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
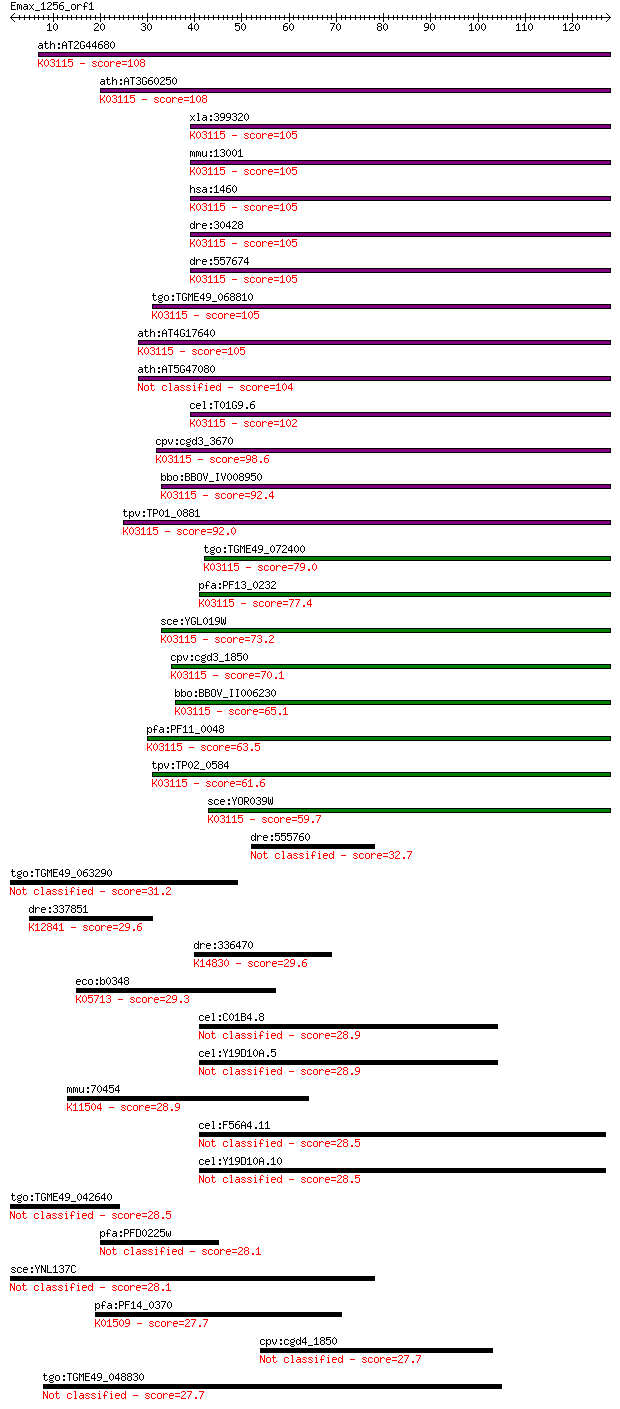
Query= Emax_1256_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
ath:AT2G44680 CKB4; CKB4 (CASEIN KINASE II BETA SUBUNIT 4); pr... 108 4e-24
ath:AT3G60250 CKB3; CKB3; protein kinase regulator/ transcript... 108 4e-24
xla:399320 csnk2b, CK2beta, ck2, ck2b; casein kinase 2, beta p... 105 4e-23
mmu:13001 Csnk2b; casein kinase 2, beta polypeptide (EC:2.7.11... 105 4e-23
hsa:1460 CSNK2B, CK2B, CK2N, CSK2B, G5A, MGC138222, MGC138224;... 105 4e-23
dre:30428 ck2b, csnk2b, wu:fa16g01; casein kinase 2 beta (EC:2... 105 5e-23
dre:557674 similar to casein kinase 2 beta subunit; CK2 beta; ... 105 5e-23
tgo:TGME49_068810 casein kinase II beta chain, putative ; K031... 105 5e-23
ath:AT4G17640 CKB2; CKB2; protein kinase CK2 regulator; K03115... 105 5e-23
ath:AT5G47080 CKB1; CKB1; protein kinase regulator 104 6e-23
cel:T01G9.6 kin-10; protein KINase family member (kin-10); K03... 102 4e-22
cpv:cgd3_3670 protein kinase CK2 regulatory subunit CK2B1 ; K0... 98.6 4e-21
bbo:BBOV_IV008950 23.m05738; casein kinase II beta 4 subunit; ... 92.4 3e-19
tpv:TP01_0881 casein kinase II beta chain; K03115 casein kinas... 92.0 4e-19
tgo:TGME49_072400 casein kinase II beta chain, putative ; K031... 79.0 3e-15
pfa:PF13_0232 PfCK2beta2; casein kinase II beta chain; K03115 ... 77.4 1e-14
sce:YGL019W CKB1; Beta regulatory subunit of casein kinase 2, ... 73.2 2e-13
cpv:cgd3_1850 casein kinase II regulatory subunit; besthit Pf ... 70.1 1e-12
bbo:BBOV_II006230 18.m06515; protein kinase CK2 regulatory sub... 65.1 6e-11
pfa:PF11_0048 CK2beta1; casein kinase II beta chain; K03115 ca... 63.5 2e-10
tpv:TP02_0584 casein kinase II subunit beta; K03115 casein kin... 61.6 7e-10
sce:YOR039W CKB2; Beta' regulatory subunit of casein kinase 2,... 59.7 2e-09
dre:555760 im:6907149; zgc:163093 32.7 0.32
tgo:TGME49_063290 rhomboid-like protease TgROM2 (EC:3.4.21.105) 31.2 0.81
dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,... 29.6 2.3
dre:336470 Gdpd1, MGC56683, Pak1ip1, fb11d05, wu:fb11d05; zgc:... 29.6 2.6
eco:b0348 mhpB, ECK0345, JW0339; 2,3-dihydroxyphenylpropionate... 29.3 3.5
cel:C01B4.8 hypothetical protein 28.9 3.8
cel:Y19D10A.5 hypothetical protein 28.9 3.8
mmu:70454 Cenpl, 2610300B10Rik, AW121806, AW550697; centromere... 28.9 4.0
cel:F56A4.11 hypothetical protein 28.5 5.2
cel:Y19D10A.10 hypothetical protein 28.5 5.2
tgo:TGME49_042640 hypothetical protein 28.5 5.9
pfa:PFD0225w conserved Plasmodium membrane protein, unknown fu... 28.1 6.7
sce:YNL137C NAM9, MNA6; Mitochondrial ribosomal component of t... 28.1 7.9
pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosi... 27.7 8.4
cpv:cgd4_1850 Ccr4p. RNAse 27.7 8.5
tgo:TGME49_048830 phospholipase C delta 1 (EC:3.1.4.11 2.7.7.48) 27.7 8.8
> ath:AT2G44680 CKB4; CKB4 (CASEIN KINASE II BETA SUBUNIT 4);
protein serine/threonine kinase; K03115 casein kinase II subunit
beta
Length=283
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 57/123 (46%), Positives = 80/123 (65%), Gaps = 3/123 (2%)
Query 7 PAAAAEGSQGSSR--DINPHLNEELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFV 64
P+ + SQ SR D+ + E SD++GS+ D D +WI WFC L+G++ F VDED++
Sbjct 59 PSTSTAKSQLHSRSPDVESDTDSEGSDVSGSEGD-DTSWISWFCNLRGNEFFCEVDEDYI 117
Query 65 RDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPSDSVSFAYIEKSAALLFGLIHARFILT 124
+DDFNL GLS QVPYY+ AL LI+D + S+ +E +A +L+GLIH R+ILT
Sbjct 118 QDDFNLCGLSGQVPYYDYALDLILDVESSNGDMFTEEQHEMVESAAEMLYGLIHVRYILT 177
Query 125 NKG 127
KG
Sbjct 178 TKG 180
> ath:AT3G60250 CKB3; CKB3; protein kinase regulator/ transcription
factor binding; K03115 casein kinase II subunit beta
Length=275
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 54/108 (50%), Positives = 73/108 (67%), Gaps = 1/108 (0%)
Query 20 DINPHLNEELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPY 79
D+ + E SD++GS+ D D +WI WFC L+G+D F VDED+++DDFNL GLS QVPY
Sbjct 68 DVESETDSEGSDVSGSEGD-DTSWISWFCNLRGNDFFCEVDEDYIQDDFNLCGLSGQVPY 126
Query 80 YEKALTLIVDGDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
Y+ AL LI+D D S+ +E +A +L+GLIH R+ILT KG
Sbjct 127 YDYALDLILDVDASNSEMFTDEQHEMVESAAEMLYGLIHVRYILTTKG 174
> xla:399320 csnk2b, CK2beta, ck2, ck2b; casein kinase 2, beta
polypeptide (EC:2.7.11.1); K03115 casein kinase II subunit
beta
Length=215
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDED+++D FNLTGL+ QVP+Y +AL +I+D + ++
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
+ IE++A +L+GLIHAR+ILTN+G
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRG 93
> mmu:13001 Csnk2b; casein kinase 2, beta polypeptide (EC:2.7.11.1);
K03115 casein kinase II subunit beta
Length=215
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDED+++D FNLTGL+ QVP+Y +AL +I+D + ++
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
+ IE++A +L+GLIHAR+ILTN+G
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRG 93
> hsa:1460 CSNK2B, CK2B, CK2N, CSK2B, G5A, MGC138222, MGC138224;
casein kinase 2, beta polypeptide (EC:2.7.11.1); K03115 casein
kinase II subunit beta
Length=215
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDED+++D FNLTGL+ QVP+Y +AL +I+D + ++
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
+ IE++A +L+GLIHAR+ILTN+G
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRG 93
> dre:30428 ck2b, csnk2b, wu:fa16g01; casein kinase 2 beta (EC:2.7.11.1);
K03115 casein kinase II subunit beta
Length=215
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDED+++D FNLTGL+ QVP+Y +AL +I+D + ++
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
+ IE++A +L+GLIHAR+ILTN+G
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRG 93
> dre:557674 similar to casein kinase 2 beta subunit; CK2 beta;
K03115 casein kinase II subunit beta
Length=215
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDED+++D FNLTGL+ QVP+Y +AL +I+D + ++
Sbjct 5 EEVSWISWFCGLRGNEFFCEVDEDYIQDKFNLTGLNEQVPHYRQALDMILDLEPDEELED 64
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
+ IE++A +L+GLIHAR+ILTN+G
Sbjct 65 NPNQSDLIEQAAEMLYGLIHARYILTNRG 93
> tgo:TGME49_068810 casein kinase II beta chain, putative ; K03115
casein kinase II subunit beta
Length=315
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 48/99 (48%), Positives = 76/99 (76%), Gaps = 2/99 (2%)
Query 31 DLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDG 90
D+TG+ ED++W++WFC LKG++ FV+VD+DF+RDDFNLTGL++QVP +++AL +++D
Sbjct 48 DMTGTSELEDMSWVEWFCTLKGNELFVVVDDDFIRDDFNLTGLASQVPLFDEALDIVLDN 107
Query 91 DISDDCPSDSVSF--AYIEKSAALLFGLIHARFILTNKG 127
+ S+D + A E++A LL+GL+HARF+ T +G
Sbjct 108 EQSEDEDDEEEQRKSAMAEQAAELLYGLVHARFLATARG 146
> ath:AT4G17640 CKB2; CKB2; protein kinase CK2 regulator; K03115
casein kinase II subunit beta
Length=282
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 52/100 (52%), Positives = 72/100 (72%), Gaps = 1/100 (1%)
Query 28 ELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLI 87
E S+++GSD ED +WI WFC L+G++ F VD+D+++DDFNL GLS+QVPYY+ AL LI
Sbjct 82 EESEVSGSD-GEDTSWISWFCNLRGNEFFCEVDDDYIQDDFNLCGLSHQVPYYDYALDLI 140
Query 88 VDGDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
+D + S IE +A +L+G+IHARFILT+KG
Sbjct 141 LDVESSHGEMFTEEQNELIESAAEMLYGMIHARFILTSKG 180
> ath:AT5G47080 CKB1; CKB1; protein kinase regulator
Length=253
Score = 104 bits (260), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 53/100 (53%), Positives = 71/100 (71%), Gaps = 1/100 (1%)
Query 28 ELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLI 87
E SD++GSD ED +WI WFC L+G++ F VD+D+++DDFNL GLS+ VPYYE AL LI
Sbjct 84 EESDVSGSD-GEDTSWISWFCNLRGNEFFCEVDDDYIQDDFNLCGLSSLVPYYEYALDLI 142
Query 88 VDGDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
+D + S IE +A +L+GLIHAR+ILT+KG
Sbjct 143 LDVESSQGEMFTEEQNELIESAAEMLYGLIHARYILTSKG 182
> cel:T01G9.6 kin-10; protein KINase family member (kin-10); K03115
casein kinase II subunit beta
Length=234
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 45/89 (50%), Positives = 69/89 (77%), Gaps = 1/89 (1%)
Query 39 EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPS 98
E+V+WI WFCGL+G++ F VDE++++D FNLTGL+ QVP Y +AL +I+D + DD
Sbjct 5 EEVSWITWFCGLRGNEFFCEVDEEYIQDRFNLTGLNEQVPKYRQALDMILDLE-PDDIED 63
Query 99 DSVSFAYIEKSAALLFGLIHARFILTNKG 127
++ + +E++A +L+GLIHAR+ILTN+G
Sbjct 64 NATNTDLVEQAAEMLYGLIHARYILTNRG 92
> cpv:cgd3_3670 protein kinase CK2 regulatory subunit CK2B1 ;
K03115 casein kinase II subunit beta
Length=252
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 51/98 (52%), Positives = 71/98 (72%), Gaps = 5/98 (5%)
Query 32 LTGSDLD--EDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVD 89
+G++LD E+ +WI+WFC LK + F+ VD++++ DDFNLTGL+ V YY+ AL +I+D
Sbjct 15 FSGNELDYEEEASWIEWFCSLKRSEFFIEVDDEYIMDDFNLTGLNEHVIYYDDALDMILD 74
Query 90 GDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
I DD D +S IE SA LL+GLIHAR+ILT+KG
Sbjct 75 R-IDDDFSEDEIS--AIESSAQLLYGLIHARYILTSKG 109
> bbo:BBOV_IV008950 23.m05738; casein kinase II beta 4 subunit;
K03115 casein kinase II subunit beta
Length=242
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 45/108 (41%), Positives = 64/108 (59%), Gaps = 13/108 (12%)
Query 33 TGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDG-- 90
+ D ++ WI+W+C LKGH+ +V VD D++RD+FNL GL QV YY A+ +I+D
Sbjct 19 SSEDQQCEIPWIEWYCSLKGHEFYVQVDADYIRDEFNLVGLQQQVSYYSHAIRMILDNYD 78
Query 91 DISDDC-----------PSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
D DC S+ I S+ LL+GLIH+RFI+T+KG
Sbjct 79 DFDYDCYEENDGDTYGPCSERAKQRIINTSSQLLYGLIHSRFIITSKG 126
> tpv:TP01_0881 casein kinase II beta chain; K03115 casein kinase
II subunit beta
Length=252
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 53/120 (44%), Positives = 73/120 (60%), Gaps = 18/120 (15%)
Query 25 LNEELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNL----------TGLS 74
++E +D T D + ++TWI+W+C LKG+ ++ VDE F+RD+FNL +GL
Sbjct 18 MSENRTD-TSDDQNSEMTWIEWYCSLKGNQYYIQVDESFIRDEFNLVGTYLPIYLFSGLQ 76
Query 75 NQVPYYEKALTLIVD---GDISDDCPSDSVSFA----YIEKSAALLFGLIHARFILTNKG 127
QV YY AL LI+D D DD +SVS I SA LL+GLIH+RFI+T+KG
Sbjct 77 YQVSYYNSALQLILDNYENDFYDDDDYESVSEKGKQHQINTSAQLLYGLIHSRFIMTSKG 136
> tgo:TGME49_072400 casein kinase II beta chain, putative ; K03115
casein kinase II subunit beta
Length=361
Score = 79.0 bits (193), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 37/86 (43%), Positives = 55/86 (63%), Gaps = 1/86 (1%)
Query 42 TWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPSDSV 101
TWI WFC L+GH+ F VDE++++D FNL GL + Y+ AL +I+ G DD D
Sbjct 85 TWISWFCALEGHECFSEVDEEYIKDTFNLFGLKPLIGNYDAALDMIL-GAAPDDEDIDEQ 143
Query 102 SFAYIEKSAALLFGLIHARFILTNKG 127
F + + A L+GLIH+R+++T +G
Sbjct 144 HFLEVYRDAMDLYGLIHSRYVITPRG 169
> pfa:PF13_0232 PfCK2beta2; casein kinase II beta chain; K03115
casein kinase II subunit beta
Length=385
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 44/104 (42%), Positives = 63/104 (60%), Gaps = 17/104 (16%)
Query 41 VTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIV------------ 88
V+WI+WFC LK + V VDEDF+RD+FNL GL +VP+++K L +I+
Sbjct 159 VSWIEWFCQLKQNLFLVEVDEDFIRDEFNLIGLQTKVPHFKKLLKIILDEDDDDDDDDDD 218
Query 89 -----DGDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
D +I+ D + E++AA L+GLIH+RFILT+KG
Sbjct 219 DYDDEDDEINRDSEEMYKNKDMHEQNAACLYGLIHSRFILTSKG 262
> sce:YGL019W CKB1; Beta regulatory subunit of casein kinase 2,
a Ser/Thr protein kinase with roles in cell growth and proliferation;
the holoenzyme also contains CKA1, CKA2 and CKB2,
the many substrates include transcription factors and all
RNA polymerases; K03115 casein kinase II subunit beta
Length=278
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 49/131 (37%), Positives = 61/131 (46%), Gaps = 38/131 (29%)
Query 33 TGSDLDEDV----TWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIV 88
TGS DED WI FC GH+ F V +F+ DDFN+T LS +VP+Y KAL LI+
Sbjct 12 TGSSDDEDSGAYDEWIPSFCSRFGHEYFCQVPTEFIEDDFNMTSLSQEVPHYRKALDLIL 71
Query 89 D--------------------------------GDISDDCPSDSVSFAYIEKSAALLFGL 116
D G + P V+ + IE +A L+GL
Sbjct 72 DLEAMSDEEEDEDDVVEEDEVDQEMQSNDGHDEGKRRNKSP--VVNKSIIEHAAEQLYGL 129
Query 117 IHARFILTNKG 127
IHARFILT G
Sbjct 130 IHARFILTKPG 140
> cpv:cgd3_1850 casein kinase II regulatory subunit; besthit Pf
23508244 ; K03115 casein kinase II subunit beta
Length=338
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 35/95 (36%), Positives = 54/95 (56%), Gaps = 2/95 (2%)
Query 35 SDLDEDVT-WIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPY-YEKALTLIVDGDI 92
SD DE W+ W+CGL+ H F+++ ++ RD FNL GL P Y+ + LI+ +I
Sbjct 23 SDSDESENGWVHWYCGLREHYFFIVIPNEYTRDAFNLYGLRQYFPKNYDSLIELILSSNI 82
Query 93 SDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
D+ + + + A L+GLIHAR+I T +G
Sbjct 83 PDEDDLADPALQDLHREAGELYGLIHARYITTPRG 117
> bbo:BBOV_II006230 18.m06515; protein kinase CK2 regulatory subunit
CK2B1; K03115 casein kinase II subunit beta
Length=245
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 57/92 (61%), Gaps = 2/92 (2%)
Query 36 DLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDD 95
D D+ + WI WF L GH+ + V+E F++D FNL GL + + Y+ A+ +I+ +++
Sbjct 18 DSDDAMKWIPWFISLDGHEFLLEVEESFIQDQFNLCGLKS-MKNYDSAMEMILGYAPTEE 76
Query 96 CPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
D+ SF + + A+ L+G+IHARFI + G
Sbjct 77 VFMDA-SFLQLYRCASDLYGMIHARFITSPMG 107
> pfa:PF11_0048 CK2beta1; casein kinase II beta chain; K03115
casein kinase II subunit beta
Length=245
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 52/98 (53%), Gaps = 4/98 (4%)
Query 30 SDLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVD 89
DL S D+ +W++WF + V VD +++ D FNL GL ++P + L++I
Sbjct 8 KDLQDSKSDKSTSWVKWFNNRALSNFLVEVDNEYITDSFNLYGLKTEIPNFNHLLSIIA- 66
Query 90 GDISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
GD +D SF+ K L+ LIHARFI T KG
Sbjct 67 GDAPEDDDDSKNSFS---KDCICLYSLIHARFITTPKG 101
> tpv:TP02_0584 casein kinase II subunit beta; K03115 casein kinase
II subunit beta
Length=242
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 33/97 (34%), Positives = 57/97 (58%), Gaps = 2/97 (2%)
Query 31 DLTGSDLDEDVTWIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDG 90
D S+ E + WI WF L+GH+ + ++E FVRD FNL+GL + + Y A+++I+
Sbjct 15 DQFSSESCETLKWIPWFTSLEGHEFLLEIEESFVRDQFNLSGLKS-LRNYNGAMSMILGP 73
Query 91 DISDDCPSDSVSFAYIEKSAALLFGLIHARFILTNKG 127
+++ D F + +A L+G++H+R+I T G
Sbjct 74 APTEEVFMDG-KFLELYMNATDLYGMLHSRYITTPIG 109
> sce:YOR039W CKB2; Beta' regulatory subunit of casein kinase
2, a Ser/Thr protein kinase with roles in cell growth and proliferation;
the holoenzyme also contains CKA1, CKA2 and CKB1,
the many substrates include transcription factors and all
RNA polymerases; K03115 casein kinase II subunit beta
Length=258
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 52/88 (59%), Gaps = 5/88 (5%)
Query 43 WIQWFCGLKGHDNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPSDSVS 102
WI F G KGH+ F VD +++ D FNL L V + + IVD D+ DD ++++
Sbjct 38 WIDLFLGRKGHEYFCDVDPEYITDRFNLMNLQKTVSKFSYVVQYIVD-DL-DDSILENMT 95
Query 103 FAYIEK---SAALLFGLIHARFILTNKG 127
A +E+ + L+GLIHAR+I+T KG
Sbjct 96 HARLEQLESDSRKLYGLIHARYIITIKG 123
> dre:555760 im:6907149; zgc:163093
Length=419
Score = 32.7 bits (73), Expect = 0.32, Method: Composition-based stats.
Identities = 14/26 (53%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 52 GHDNFVLVDEDFVRDDFNLTGLSNQV 77
GH F +V + F RD+ N GLSNQV
Sbjct 284 GHTQFHIVPDRFCRDNMNKRGLSNQV 309
> tgo:TGME49_063290 rhomboid-like protease TgROM2 (EC:3.4.21.105)
Length=283
Score = 31.2 bits (69), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 7/48 (14%)
Query 1 SHARGGPAAAAEGSQGSSRDINPHLNEELSDLTGSDLDEDVTWIQWFC 48
S RG A EG QG S L +G+ E+V+WIQW C
Sbjct 13 SPPRGSSALEGEGRQGGS-------PLPLFFPSGTQSGENVSWIQWLC 53
> dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,
xx:tdsubc_2h12, zgc:55518; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic
reticulum protein
Length=909
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 5 GGPAAAAEGSQGSSRDINPHLNEELS 30
G P++ EGSQ S D NP L+ E+S
Sbjct 374 GPPSSEYEGSQNRSSDSNPALSPEMS 399
> dre:336470 Gdpd1, MGC56683, Pak1ip1, fb11d05, wu:fb11d05; zgc:56683;
K14830 protein MAK11
Length=368
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 40 DVTWIQWFCGLKGHDNFVLVDEDFVRDDF 68
DVT + C K H+N V E F++DDF
Sbjct 236 DVTSQKCVCEFKAHENRVKAIESFMKDDF 264
> eco:b0348 mhpB, ECK0345, JW0339; 2,3-dihydroxyphenylpropionate
1,2-dioxygenase (EC:1.13.11.-); K05713 2,3-dihydroxyphenylpropionate
1,2-dioxygenase [EC:1.13.11.16]
Length=314
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 15 QGSSRDINPHLNEELSDLTGSDLDEDVTWIQWFCGLKGHDNF 56
QG ++++ NEELS + G E TW+ F + N+
Sbjct 247 QGRIQELDAVSNEELSAIAGKSTHEIKTWVAAFAAISAFGNW 288
> cel:C01B4.8 hypothetical protein
Length=478
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 31/69 (44%), Gaps = 6/69 (8%)
Query 41 VTWIQWFCGLKGHDNFVLVDEDFVRDDFNL----TGLSNQVPYYEKALTLIVDGDISDDC 96
VTW+ G G VL ++R+ N TG ++ +P+ A + G +SD C
Sbjct 270 VTWMSNIGGNLGFLTLVLYGPTYLREVLNFEVRGTGFASALPFLLSAAVKSIAGQLSDRC 329
Query 97 P--SDSVSF 103
S+ V F
Sbjct 330 DFVSERVRF 338
> cel:Y19D10A.5 hypothetical protein
Length=478
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 31/69 (44%), Gaps = 6/69 (8%)
Query 41 VTWIQWFCGLKGHDNFVLVDEDFVRDDFNL----TGLSNQVPYYEKALTLIVDGDISDDC 96
VTW+ G G VL ++R+ N TG ++ +P+ A + G +SD C
Sbjct 270 VTWMSNIGGNLGFLTLVLYGPTYLREVLNFEVRGTGFASALPFLLSAAVKSIAGQLSDRC 329
Query 97 P--SDSVSF 103
S+ V F
Sbjct 330 DFVSERVRF 338
> mmu:70454 Cenpl, 2610300B10Rik, AW121806, AW550697; centromere
protein L; K11504 centromere protein L
Length=345
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 24/53 (45%), Gaps = 2/53 (3%)
Query 13 GSQGSSRDINPHLNEELSDLTGSD--LDEDVTWIQWFCGLKGHDNFVLVDEDF 63
G +G+ RD L + LS S ++ V W WFC + G V EDF
Sbjct 127 GVKGTQRDHEAFLVQILSKSQSSREHREDKVLWTGWFCCVFGESLQETVSEDF 179
> cel:F56A4.11 hypothetical protein
Length=746
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query 41 VTWIQWFCGLKGHDNFVLVDEDFVRD----DFNLTGLSNQVPYYEKALTLIVDGDISDDC 96
+TWI F G G + L ++R+ D TG + +P+ A+ G ISD
Sbjct 547 ITWITCFGGFVGLYSLSLYGPTYLREILKFDVRETGFLSALPFILSAIIKFAAGKISDKL 606
Query 97 P--SDSVSFAYIEKSAALLF--GLIHARFILTNK 126
S+ F + ++ + F GL+ F TN+
Sbjct 607 SFLSEKSRFVFFAATSQIGFVAGLLVMAFT-TNR 639
> cel:Y19D10A.10 hypothetical protein
Length=703
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query 41 VTWIQWFCGLKGHDNFVLVDEDFVRD----DFNLTGLSNQVPYYEKALTLIVDGDISDDC 96
+TWI F G G + L ++R+ D TG + +P+ A+ G ISD
Sbjct 504 ITWITCFGGFVGLYSLSLYGPTYLREILKFDVRETGFLSALPFILSAIIKFAAGKISDKL 563
Query 97 P--SDSVSFAYIEKSAALLF--GLIHARFILTNK 126
S+ F + ++ + F GL+ F TN+
Sbjct 564 SFLSEKSRFVFFAATSQIGFVAGLLVMAFT-TNR 596
> tgo:TGME49_042640 hypothetical protein
Length=3225
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 1 SHARGGPAAAAEGSQGSSRDINP 23
S RGG AA+EGS G + D++P
Sbjct 2763 SVGRGGAVAASEGSVGETVDVSP 2785
> pfa:PFD0225w conserved Plasmodium membrane protein, unknown
function
Length=4138
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 18/25 (72%), Gaps = 2/25 (8%)
Query 20 DINPHLNEELSDLTGSDLDEDVTWI 44
D+N H+N +++ DLDED+TW+
Sbjct 1326 DVNNHVNHDVN--ADDDLDEDMTWM 1348
> sce:YNL137C NAM9, MNA6; Mitochondrial ribosomal component of
the small subunit
Length=486
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 32/81 (39%), Gaps = 7/81 (8%)
Query 1 SHARGGPAAAAEGSQGSSRDINPHLNEELSDLTGSDLDEDVTWIQWFCGLKGHDNFVLV- 59
S R G AE + +++I H +SD++ L D W + LK HD L
Sbjct 334 SALRSGKRIIAESVKLWTKNITDHFKTRMSDISDGSLTFDPKWAK---NLKYHDPIKLSE 390
Query 60 ---DEDFVRDDFNLTGLSNQV 77
DE R NL N V
Sbjct 391 LEGDEPKARKLINLPWQKNYV 411
> pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2472
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Query 19 RDINPHLNEELSDLTGSDLDEDVTWIQW-FCGLKGHDNFVLVDEDFVRDDFNL 70
++I HLN E+S T ++++ + W+++ + ++ N L D D + +D NL
Sbjct 869 KNIENHLNAEISIGTIKNIEDGIKWLEYTYLFIRMKKNPYLYDVD-INNDLNL 920
> cpv:cgd4_1850 Ccr4p. RNAse
Length=689
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 54 DNFVLVDEDFVRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPSDSVS 102
D LV+E+ D FN+ L+N +PY ++A ++ S PSD +S
Sbjct 630 DYIYLVEEEAFSDKFNIF-LNNFLPYIDEAALSPINTLPSPQYPSDHIS 677
> tgo:TGME49_048830 phospholipase C delta 1 (EC:3.1.4.11 2.7.7.48)
Length=1097
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 25/101 (24%), Positives = 42/101 (41%), Gaps = 9/101 (8%)
Query 8 AAAAEGSQGSSRDINPHLNEELSDLTGSD----LDEDVTWIQWFCGLKGHDNFVLVDEDF 63
A ++G QGS ++ + + L+D+ + L E ++W C NF E F
Sbjct 100 ALPSQGRQGSGAVVSQRIKDLLADVDTREALQTLAEGSFLLKW-C----KTNFKKPHERF 154
Query 64 VRDDFNLTGLSNQVPYYEKALTLIVDGDISDDCPSDSVSFA 104
D N L + P +++T I GD+ P + F
Sbjct 155 FYVDANAMALKWRSPKKPESVTTIPVGDVQRIIPGEDTDFG 195
Lambda K H
0.317 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40