bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1313_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
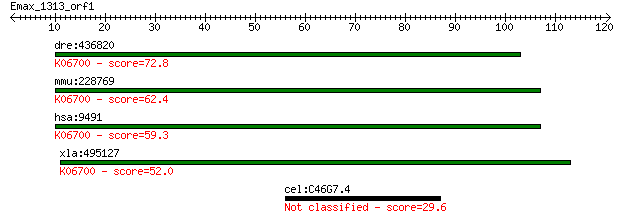
dre:436820 psmf1, zgc:92785; proteasome (prosome, macropain) i... 72.8 3e-13
mmu:228769 Psmf1, AW048666, BC012260, MGC18784, PI31; proteaso... 62.4 3e-10
hsa:9491 PSMF1, PI31; proteasome (prosome, macropain) inhibito... 59.3 3e-09
xla:495127 MGC130674; hypothetical LOC495127; K06700 proteasom... 52.0 4e-07
cel:C46G7.4 pqn-22; Prion-like-(Q/N-rich)-domain-bearing prote... 29.6 2.2
> dre:436820 psmf1, zgc:92785; proteasome (prosome, macropain)
inhibitor subunit 1; K06700 proteasome inhibitor subunit 1
(PI31)
Length=272
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 57/93 (61%), Gaps = 3/93 (3%)
Query 10 MKGFQIFFETVKSDLTSKLDVIVSVIHFLLVDDDFVCVGVGEKFTDADATGGSELLPSGW 69
M G ++ F V + LT D ++ +H+ +V + C+G+G++ D + +ELLPSGW
Sbjct 1 MAGLELLFNCVSNSLTCSQDALICFVHWEIVKSGYKCMGIGDEPKDGEKK--TELLPSGW 58
Query 70 NSNQAIYSLKYRSRHNDKISLLLKGIMAGDLLI 102
N ++ +Y+L+YRS ++DK +LLLK LI
Sbjct 59 NESKELYALRYRS-NDDKSNLLLKAFTVDSSLI 90
> mmu:228769 Psmf1, AW048666, BC012260, MGC18784, PI31; proteasome
(prosome, macropain) inhibitor subunit 1; K06700 proteasome
inhibitor subunit 1 (PI31)
Length=271
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 56/97 (57%), Gaps = 3/97 (3%)
Query 10 MKGFQIFFETVKSDLTSKLDVIVSVIHFLLVDDDFVCVGVGEKFTDADATGGSELLPSGW 69
M G ++ F + ++ D +V +H+ +V + + +G G++ +D SELLP+ W
Sbjct 1 MAGLEVLFASAAPSMSCPQDALVCFLHWEVVTNGYYALGTGDQPGPSDKK--SELLPAKW 58
Query 70 NSNQAIYSLKYRSRHNDKISLLLKGIMAGDLLIVSAL 106
NSN+ +Y+L+Y S+ + LLLK + + +I++ L
Sbjct 59 NSNKELYALRYESKDGAR-KLLLKAVSVENGMIINVL 94
> hsa:9491 PSMF1, PI31; proteasome (prosome, macropain) inhibitor
subunit 1 (PI31); K06700 proteasome inhibitor subunit 1
(PI31)
Length=271
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query 10 MKGFQIFFETVKSDLTSKLDVIVSVIHFLLVDDDFVCVGVGEKFTDADATGGSELLPSGW 69
M G ++ F + +T + D +V +H+ +V + +GVG++ D SELLP+GW
Sbjct 1 MAGLEVLFASAAPAITCRQDALVCFLHWEVVTHGYFGLGVGDQPGPNDKK--SELLPAGW 58
Query 70 NSNQAIYSLKYRSRHNDKISLLLKGIMAGDLLIVSAL 106
N+N+ +Y L+Y + + LL+K I +I++ L
Sbjct 59 NNNKDLYVLRYEYKDGSR-KLLVKAITVESSMILNVL 94
> xla:495127 MGC130674; hypothetical LOC495127; K06700 proteasome
inhibitor subunit 1 (PI31)
Length=263
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 54/102 (52%), Gaps = 5/102 (4%)
Query 11 KGFQIFFETVKSDLTSKLDVIVSVIHFLLVDDDFVCVGVGEKFTDADATGGSELLPSGWN 70
G ++ F S+++ D ++ IH+ L+ C+G GE+ A TG SE LP GW
Sbjct 4 PGLELLFSLFSSEISRPTDSLICFIHWELICRGLRCLGRGEE-AGAQETG-SERLPVGWA 61
Query 71 SNQAIYSLKYRSRHNDKISLLLKGIMAGDLLIVSALPMNSSN 112
N+ +Y+L Y S ++ +LLK + +IV+ + M++
Sbjct 62 ENKDLYTLCYGSPNS---QILLKALTVEGTVIVNIMDMHTEK 100
> cel:C46G7.4 pqn-22; Prion-like-(Q/N-rich)-domain-bearing protein
family member (pqn-22)
Length=1175
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 56 ADATGGSELLPSGWNSNQAIYSLKYRSRHND 86
+D + LL SG+N+NQ Y L ++ ++N+
Sbjct 639 SDHSNAQRLLNSGYNTNQVDYVLGHKDKYNE 669
Lambda K H
0.320 0.136 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40