bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1361_orf1
Length=165
Score E
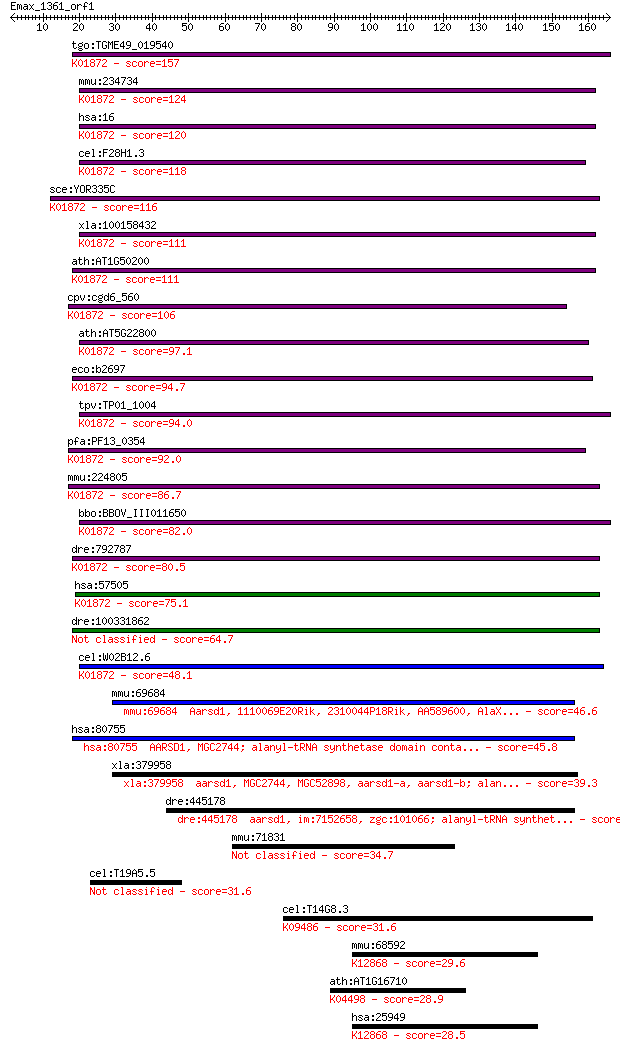
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019540 alanyl-tRNA synthetase, putative (EC:6.1.1.7... 157 1e-38
mmu:234734 Aars, AI316495, C76919, MGC37368, sti; alanyl-tRNA ... 124 2e-28
hsa:16 AARS, CMT2N; alanyl-tRNA synthetase (EC:6.1.1.7); K0187... 120 2e-27
cel:F28H1.3 ars-2; Alanyl tRNA Synthetase family member (ars-2... 118 1e-26
sce:YOR335C ALA1, CDC64; Ala1p (EC:6.1.1.7); K01872 alanyl-tRN... 116 4e-26
xla:100158432 aars; alanyl-tRNA synthetase (EC:6.1.1.7); K0187... 111 9e-25
ath:AT1G50200 ALATS; ALATS (ALANYL-TRNA SYNTHETASE); ATP bindi... 111 1e-24
cpv:cgd6_560 alanyl-tRNA synthetase (with HxxxH domain) ; K018... 106 3e-23
ath:AT5G22800 EMB1030 (EMBRYO DEFECTIVE 1030); ATP binding / a... 97.1 3e-20
eco:b2697 alaS, act, ECK2692, JW2667, lovB; alanyl-tRNA synthe... 94.7 1e-19
tpv:TP01_1004 alanyl-tRNA synthetase; K01872 alanyl-tRNA synth... 94.0 2e-19
pfa:PF13_0354 alanine--tRNA ligase, putative (EC:6.1.1.7); K01... 92.0 9e-19
mmu:224805 Aars2, Aarsl, AlaRS, Gm89, MGC69820; alanyl-tRNA sy... 86.7 3e-17
bbo:BBOV_III011650 17.m07993; alanyl-tRNA synthetase family pr... 82.0 7e-16
dre:792787 im:7141191; si:dkey-240e12.1; K01872 alanyl-tRNA sy... 80.5 2e-15
hsa:57505 AARS2, AARSL, KIAA1270, MT-ALARS, MTALARS, bA444E17.... 75.1 1e-13
dre:100331862 Alanyl tRNA Synthetase family member (ars-2)-like 64.7 1e-10
cel:W02B12.6 ars-1; Alanyl tRNA Synthetase family member (ars-... 48.1 1e-05
mmu:69684 Aarsd1, 1110069E20Rik, 2310044P18Rik, AA589600, AlaX... 46.6 4e-05
hsa:80755 AARSD1, MGC2744; alanyl-tRNA synthetase domain conta... 45.8 7e-05
xla:379958 aarsd1, MGC2744, MGC52898, aarsd1-a, aarsd1-b; alan... 39.3 0.005
dre:445178 aarsd1, im:7152658, zgc:101066; alanyl-tRNA synthet... 38.1 0.012
mmu:71831 1700007B14Rik, MGC143580, MGC143581; RIKEN cDNA 1700... 34.7 0.14
cel:T19A5.5 nhr-219; Nuclear Hormone Receptor family member (n... 31.6 1.3
cel:T14G8.3 hypothetical protein; K09486 hypoxia up-regulated 1 31.6
mmu:68592 Syf2, 1110018L13Rik, Cbpin, D4Bwg1551e, Gcipip, mp29... 29.6 5.0
ath:AT1G16710 HAC12; HAC12 (histone acetyltransferase of the C... 28.9 9.1
hsa:25949 SYF2, CBPIN, DKFZp564O2082, NTC31, P29, fSAP29; SYF2... 28.5 9.7
> tgo:TGME49_019540 alanyl-tRNA synthetase, putative (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1037
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 84/148 (56%), Positives = 110/148 (74%), Gaps = 0/148 (0%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
I+QKGSLVE +RLRFD+ + PL N+ I A+E V+ IA + PV+ +PL+ AM++P
Sbjct 695 IQQKGSLVEVKRLRFDYAGEKPLENEDIIAVENVVKDCIAQARPVHYKEVPLEQAMKIPG 754
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L AVFGEVYPDPVRVVS+G D + L + ++ + AS+ELCGGTHL NASQL DF +
Sbjct 755 LRAVFGEVYPDPVRVVSVGEDIESLVALADSSQPETPFASIELCGGTHLHNASQLSDFCI 814
Query 138 ISEESIAKGIRRIVAVAGEAAVSARSEL 165
+SEES+AKG+RR+VA AG+AA AR EL
Sbjct 815 LSEESVAKGVRRVVAAAGDAAFEARKEL 842
> mmu:234734 Aars, AI316495, C76919, MGC37368, sti; alanyl-tRNA
synthetase (EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 72/142 (50%), Positives = 85/142 (59%), Gaps = 2/142 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGSLV RLRFDF ++ QQI+ E V I ++ PV T PL A + L
Sbjct 624 QKGSLVAPDRLRFDFTAKGAMSTQQIKKAEEIVNGMIEAAKPVYTQDCPLAAAKAIQGLR 683
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
AVF E YPDPVRVVSIG EL + + G SVE CGGTHL N+S FV+++
Sbjct 684 AVFDETYPDPVRVVSIGVPVSEL--LDDPCGPAGSLTSVEFCGGTHLRNSSHAGAFVIVT 741
Query 140 EESIAKGIRRIVAVAGEAAVSA 161
EE+IAKGIRRIVAV G A A
Sbjct 742 EEAIAKGIRRIVAVTGAEAQKA 763
> hsa:16 AARS, CMT2N; alanyl-tRNA synthetase (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 70/142 (49%), Positives = 83/142 (58%), Gaps = 2/142 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGSLV RLRFDF ++ QQI+ E I ++ V T PL A + L
Sbjct 624 QKGSLVAPDRLRFDFTAKGAMSTQQIKKAEEIANEMIEAAKAVYTQDCPLAAAKAIQGLR 683
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
AVF E YPDPVRVVSIG EL + + G SVE CGGTHL N+S FV+++
Sbjct 684 AVFDETYPDPVRVVSIGVPVSEL--LDDPSGPAGSLTSVEFCGGTHLRNSSHAGAFVIVT 741
Query 140 EESIAKGIRRIVAVAGEAAVSA 161
EE+IAKGIRRIVAV G A A
Sbjct 742 EEAIAKGIRRIVAVTGAEAQKA 763
> cel:F28H1.3 ars-2; Alanyl tRNA Synthetase family member (ars-2);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 65/139 (46%), Positives = 85/139 (61%), Gaps = 2/139 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGSLV R+RFDF A +T QQ++ E Y Q+ I + V PL A ++ L
Sbjct 625 QKGSLVAPDRMRFDFTNKAGMTVQQVKKAEEYAQQLIDTKGQVYAKNSPLGEAKKVKGLR 684
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
A+F E YPDPVRVV++G ++L +Q + N +VE CGGTHL N S + V+ S
Sbjct 685 AMFDETYPDPVRVVAVGTPVEQL--LQNPDAEEGQNTTVEFCGGTHLQNVSHIGRIVIAS 742
Query 140 EESIAKGIRRIVAVAGEAA 158
EE+IAKGIRRIVA+ G A
Sbjct 743 EEAIAKGIRRIVALTGPEA 761
> sce:YOR335C ALA1, CDC64; Ala1p (EC:6.1.1.7); K01872 alanyl-tRNA
synthetase [EC:6.1.1.7]
Length=958
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 62/151 (41%), Positives = 87/151 (57%), Gaps = 2/151 (1%)
Query 12 ETERRTIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKT 71
ET + QKGSLV +LRFDF ++N++++ +E I +L V +PL
Sbjct 612 ETLGNDVDQKGSLVAPEKLRFDFSHKKAVSNEELKKVEDICNEQIKENLQVFYKEIPLDL 671
Query 72 AMELPALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQ 131
A + + AVFGE YPDPVRVVS+G +EL + + S+E CGGTH++
Sbjct 672 AKSIDGVRAVFGETYPDPVRVVSVGKPIEEL--LANPANEEWTKYSIEFCGGTHVNKTGD 729
Query 132 LEDFVVISEESIAKGIRRIVAVAGEAAVSAR 162
++ FV++ E IAKGIRRIVAV G A A+
Sbjct 730 IKYFVILEESGIAKGIRRIVAVTGTEAFEAQ 760
> xla:100158432 aars; alanyl-tRNA synthetase (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=968
Score = 111 bits (278), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 65/142 (45%), Positives = 80/142 (56%), Gaps = 2/142 (1%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
Q+GSLV RLRFDF +T Q+I E I + V PL TA + L
Sbjct 624 QRGSLVAPDRLRFDFTAKGAMTTQEIRKTEEIANDIIKENKVVYALDCPLATAKAIQGLR 683
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
AVF E YPDPVRVVSIG ++L + + SVE CGGTHL N+ F++++
Sbjct 684 AVFDETYPDPVRVVSIGIPVEDL--LADPSSPAGSVTSVEFCGGTHLKNSGHAGPFIIVT 741
Query 140 EESIAKGIRRIVAVAGEAAVSA 161
EE+IAKGIRRIVAV G A A
Sbjct 742 EEAIAKGIRRIVAVTGAEAQKA 763
> ath:AT1G50200 ALATS; ALATS (ALANYL-TRNA SYNTHETASE); ATP binding
/ alanine-tRNA ligase/ ligase, forming aminoacyl-tRNA and
related compounds / nucleic acid binding / nucleotide binding
(EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1003
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 63/144 (43%), Positives = 84/144 (58%), Gaps = 2/144 (1%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
I QKGS+V +LRFDF P+ + + IE V + I L V + L A +
Sbjct 664 IDQKGSIVLPEKLRFDFSHGKPVDPEDLRRIESIVNKQIKDELDVFSKEAVLSEAKRIKG 723
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L AVFGEVYPDPVRVVSIG ++L + + + + S E CGGTH++N + + F +
Sbjct 724 LRAVFGEVYPDPVRVVSIGRKVEDL--LADPENNEWSLLSSEFCGGTHITNTREAKAFAL 781
Query 138 ISEESIAKGIRRIVAVAGEAAVSA 161
+SEE IAKGIRR+ AV E A A
Sbjct 782 LSEEGIAKGIRRVTAVTTECAFDA 805
> cpv:cgd6_560 alanyl-tRNA synthetase (with HxxxH domain) ; K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1003
Score = 106 bits (265), Expect = 3e-23, Method: Composition-based stats.
Identities = 64/137 (46%), Positives = 83/137 (60%), Gaps = 1/137 (0%)
Query 17 TIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELP 76
T Q+GS+V+ +LRFDF PLT QIE IE + + I V + L A ++P
Sbjct 655 TCDQRGSVVDPNKLRFDFSSQKPLTLDQIEEIEKMINQIIQEKQTVYCQTVELSIAKQIP 714
Query 77 ALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFV 136
+ A+FGE YPDPVRV+S+ L Q T D+ S+E CGGTH+ N S++ F
Sbjct 715 NIRAIFGETYPDPVRVLSVSKSVDSLINSQ-TDNNDQEKVSIEFCGGTHVENTSEIISFS 773
Query 137 VISEESIAKGIRRIVAV 153
+ISEE IAKGIRRIVAV
Sbjct 774 IISEEGIAKGIRRIVAV 790
> ath:AT5G22800 EMB1030 (EMBRYO DEFECTIVE 1030); ATP binding /
alanine-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleic acid binding / nucleotide binding (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=978
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 59/141 (41%), Positives = 80/141 (56%), Gaps = 20/141 (14%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
Q GSLV RLRFDF+++ L + ++E IE + R I + + T VLPL A A+
Sbjct 675 QAGSLVAFDRLRFDFNFNRSLHDNELEEIECLINRWIGDATRLETKVLPLADAKRAGAI- 733
Query 80 AVFGEVYP-DPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVI 138
A+FGE Y + VRVV + S+ELCGGTH+ N +++ F +I
Sbjct 734 AMFGEKYDENEVRVVEVP------------------GVSMELCGGTHVGNTAEIRAFKII 775
Query 139 SEESIAKGIRRIVAVAGEAAV 159
SE+ IA GIRRI AVAGEA +
Sbjct 776 SEQGIASGIRRIEAVAGEAFI 796
> eco:b2697 alaS, act, ECK2692, JW2667, lovB; alanyl-tRNA synthetase
(EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=876
Score = 94.7 bits (234), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 79/143 (55%), Gaps = 19/143 (13%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
+ QKGSLV ++ LRFDF + + ++I A+E V I +LP+ T ++ L+ A A
Sbjct 582 VSQKGSLVNDKVLRFDFSHNEAMKPEEIRAVEDLVNTQIRRNLPIETNIMDLEAAKAKGA 641
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
+ A+FGE Y + VRV+S+G + S ELCGGTH S + F +
Sbjct 642 M-ALFGEKYDERVRVLSMG------------------DFSTELCGGTHASRTGDIGLFRI 682
Query 138 ISEESIAKGIRRIVAVAGEAAVS 160
ISE A G+RRI AV GE A++
Sbjct 683 ISESGTAAGVRRIEAVTGEGAIA 705
> tpv:TP01_1004 alanyl-tRNA synthetase; K01872 alanyl-tRNA synthetase
[EC:6.1.1.7]
Length=1011
Score = 94.0 bits (232), Expect = 2e-19, Method: Composition-based stats.
Identities = 47/146 (32%), Positives = 81/146 (55%), Gaps = 9/146 (6%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
Q+GS ++E +L+FD+ + L + I +E + + S P+ T + K A+E+P +
Sbjct 672 QRGSQLDEEKLKFDYSYGQQLPDNLIRQVEQRMMEFVRSGAPLRTTEIEYKKALEIPGIR 731
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
A F E YP+ VRVV++ +++ + S E+CGGTHL+N ++D V+
Sbjct 732 ANFDEKYPEVVRVVNVQMGSEDFS---------GEHTSTEVCGGTHLTNTKTIQDVAVVG 782
Query 140 EESIAKGIRRIVAVAGEAAVSARSEL 165
EE I+KG+RR+ V + +R +L
Sbjct 783 EEGISKGVRRLTIVTNDQCRRSRIDL 808
> pfa:PF13_0354 alanine--tRNA ligase, putative (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=1408
Score = 92.0 bits (227), Expect = 9e-19, Method: Composition-based stats.
Identities = 55/143 (38%), Positives = 76/143 (53%), Gaps = 4/143 (2%)
Query 17 TIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELP 76
T +QKGSLV++ +LRFDF + + I IE + + I L V + L + ++
Sbjct 1071 TCEQKGSLVDDEKLRFDFSFIENINIDAITKIENEINKLIKEELNVTVKTMDLDESKKIK 1130
Query 77 ALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINA-SVELCGGTHLSNASQLEDF 135
+ A+F E Y D V VV I K++ I D S+ELCGGTH+ N + F
Sbjct 1131 GIRAIFEEDYADKVNVVFI---NKDVNNILNNMNIDYTYLHSIELCGGTHIGNTKLIRKF 1187
Query 136 VVISEESIAKGIRRIVAVAGEAA 158
+V SEESI KGI RI AV + A
Sbjct 1188 LVTSEESIGKGIYRITAVTNKKA 1210
> mmu:224805 Aars2, Aarsl, AlaRS, Gm89, MGC69820; alanyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.7); K01872
alanyl-tRNA synthetase [EC:6.1.1.7]
Length=980
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 57/154 (37%), Positives = 81/154 (52%), Gaps = 20/154 (12%)
Query 17 TIKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELP 76
T +Q+GS + RLRFD LT +Q+ +E YVQ + PV +PL +P
Sbjct 644 TTEQRGSHLNPERLRFDVATQTLLTTEQLRTVESYVQEVVGQDKPVFMEEVPLAHTARIP 703
Query 77 ALCAVFGEVYPDPVRVV--------SIGPDTKELKTIQETQGADKINASVELCGGTHLSN 128
L ++ EVYPDPVRVV ++GP ++ ++ SVELC GTHL +
Sbjct 704 GLRSL-DEVYPDPVRVVSVGVPVAHALGPASQA-----------AMHTSVELCCGTHLLS 751
Query 129 ASQLEDFVVISEESIAKGIRRIVAVAGEAAVSAR 162
+ D V+I E + KGI R++A+ GE A AR
Sbjct 752 TGAVGDLVIIGERQLVKGITRLLAITGEQAQQAR 785
> bbo:BBOV_III011650 17.m07993; alanyl-tRNA synthetase family
protein (EC:6.1.1.7); K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=978
Score = 82.0 bits (201), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 45/146 (30%), Positives = 83/146 (56%), Gaps = 9/146 (6%)
Query 20 QKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALC 79
QKGS ++E +L+FD + + ++ ++ IE +Q I ++ + + K A+E+P +
Sbjct 641 QKGSQLDEEKLKFDIAANKQMPDETLQRIEDRMQEIIDANWKIEVREIDFKDAVEIPGIR 700
Query 80 AVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVIS 139
A F +VYP+ VRVV++ D + + D SVE+CGGTH+ L+ +++
Sbjct 701 ANFTDVYPEVVRVVAMTKDGQPI---------DGKAYSVEICGGTHVPFTGVLKSAIIVG 751
Query 140 EESIAKGIRRIVAVAGEAAVSARSEL 165
EES++KG+RR+ E + +++ L
Sbjct 752 EESVSKGVRRLTLATNEKSEDSKAIL 777
> dre:792787 im:7141191; si:dkey-240e12.1; K01872 alanyl-tRNA
synthetase [EC:6.1.1.7]
Length=1004
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 56/145 (38%), Positives = 79/145 (54%), Gaps = 6/145 (4%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
+ Q+GS RLRFDF LT +++ +E V I + V+ +PL +A ++
Sbjct 660 VSQRGSHCSANRLRFDFSVKGTLTVSELQKVEELVLNIIRQNADVHVEEVPLSSAKQIAG 719
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L V E+YPDPVRVVS+ +L + + SVELC GTHL + DFVV
Sbjct 720 LRTV-DELYPDPVRVVSVAVPVSDLLASERAE-----QTSVELCCGTHLLRTGAIRDFVV 773
Query 138 ISEESIAKGIRRIVAVAGEAAVSAR 162
+SE + KG+ RIVA G+ A+ AR
Sbjct 774 VSERQMMKGVCRIVAFTGDDAIKAR 798
> hsa:57505 AARS2, AARSL, KIAA1270, MT-ALARS, MTALARS, bA444E17.1;
alanyl-tRNA synthetase 2, mitochondrial (putative) (EC:6.1.1.7);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=985
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 53/144 (36%), Positives = 77/144 (53%), Gaps = 4/144 (2%)
Query 19 KQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPAL 78
+Q+GS + +LR D PLT +Q+ A+E VQ A+ V +PL ++P L
Sbjct 651 EQQGSHLNPEQLRLDVTTQTPLTPEQLRAVENTVQEAVGQDEAVYMEEVPLALTAQVPGL 710
Query 79 CAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVI 138
++ EVYPDPVRVVS+ + + + SVELC GTHL + D V+I
Sbjct 711 RSL-DEVYPDPVRVVSV---GVPVAHALDPASQAALQTSVELCCGTHLLRTGAVGDLVII 766
Query 139 SEESIAKGIRRIVAVAGEAAVSAR 162
+ ++KG R++AV GE A AR
Sbjct 767 GDRQLSKGTTRLLAVTGEQAQQAR 790
> dre:100331862 Alanyl tRNA Synthetase family member (ars-2)-like
Length=796
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 63/145 (43%), Gaps = 35/145 (24%)
Query 18 IKQKGSLVEERRLRFDFDWDAPLTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPA 77
+ Q+GS RLRFDF LT +++ +E V I + V+ +PL +A ++
Sbjct 481 VSQRGSHCSANRLRFDFSVKGSLTVSELQKVEELVLNIIRQNADVHVEEVPLSSAKQIAG 540
Query 78 LCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVV 137
L V E+YPDPV HL + DFVV
Sbjct 541 LRTV-DELYPDPV----------------------------------HLLRTGAIRDFVV 565
Query 138 ISEESIAKGIRRIVAVAGEAAVSAR 162
+SE + KG+ RIVA G+ A+ AR
Sbjct 566 VSERQMMKGVCRIVAFTGDDAIKAR 590
> cel:W02B12.6 ars-1; Alanyl tRNA Synthetase family member (ars-1);
K01872 alanyl-tRNA synthetase [EC:6.1.1.7]
Length=793
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 41/151 (27%), Positives = 73/151 (48%), Gaps = 22/151 (14%)
Query 20 QKGSLVEERRLRFDFD-WDAPLTNQQ----IEAIELYVQRAIASSLPVNTAVLPLKTAME 74
QKGS V+ R RFD+ D L+ +Q + E+ ++ I + L+ A +
Sbjct 613 QKGSSVDCDRFRFDYSTGDEDLSKEQRTELLIKCEMKMREFIQNGGFTEIIETSLEEAKK 672
Query 75 LPALCAVFGE--VYPDPVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQL 132
+ L + E + VRVV++G GAD VE C GTH+ + +
Sbjct 673 IENLQSDVKEDRIGGASVRVVALGS------------GAD---VPVECCSGTHIHDVRVI 717
Query 133 EDFVVISEESIAKGIRRIVAVAGEAAVSARS 163
+D ++S++S+ + +RRI+ + G+ A + R+
Sbjct 718 DDVAIMSDKSMGQRLRRIIVLTGKEAAACRN 748
> mmu:69684 Aarsd1, 1110069E20Rik, 2310044P18Rik, AA589600, AlaX,
AlaXp-II; alanyl-tRNA synthetase domain containing 1; K07050
Length=412
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 41/132 (31%), Positives = 60/132 (45%), Gaps = 27/132 (20%)
Query 29 RLRFDFDWDAP-LTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYP 87
RLR + D+P +T +Q+ AIE V + I LPV+ L L P + V G P
Sbjct 134 RLRSVIELDSPSVTAEQVAAIEQSVNQKIRDRLPVSVRELSLDD----PEVEQVRGRGLP 189
Query 88 D----PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESI 143
D P+RVV+I +G D +C GTH+SN S L+ ++ E
Sbjct 190 DDHAGPIRVVTI-------------EGVDS-----NMCCGTHVSNLSDLQVIKILGTEKG 231
Query 144 AKGIRRIVAVAG 155
K ++ +AG
Sbjct 232 KKNKSNLIFLAG 243
> hsa:80755 AARSD1, MGC2744; alanyl-tRNA synthetase domain containing
1; K07050
Length=586
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 61/143 (42%), Gaps = 27/143 (18%)
Query 18 IKQKGSLVEERRLRFDFDWDAP-LTNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELP 76
K K + E R R + D P +T +Q+ AIE V I LPVN L L P
Sbjct 297 FKLKTTSWELGRFRSAIELDTPSMTAEQVAAIEQSVNEKIRDRLPVNVRELSLDD----P 352
Query 77 ALCAVFGEVYPD----PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQL 132
+ V G PD P+RVV+I +G D +C GTH+SN S L
Sbjct 353 EVEQVSGRGLPDDHAGPIRVVNI-------------EGVDS-----NMCCGTHVSNLSDL 394
Query 133 EDFVVISEESIAKGIRRIVAVAG 155
+ ++ E K ++ ++G
Sbjct 395 QVIKILGTEKGKKNRTNLIFLSG 417
> xla:379958 aarsd1, MGC2744, MGC52898, aarsd1-a, aarsd1-b; alanyl-tRNA
synthetase domain containing 1; K07050
Length=412
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 57/133 (42%), Gaps = 27/133 (20%)
Query 29 RLRFDFDWDAPL-TNQQIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYP 87
R R + D PL T +Q+EAIE + I +PV+ ++ ++ P V P
Sbjct 133 RQRSVIELDTPLVTAEQVEAIEKVANQKIREHVPVHVRLI----TVDDPEFDMVRSRGLP 188
Query 88 D----PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESI 143
D PVR++ I +G D +C GTH+ N S L+ ++ E
Sbjct 189 DDHAGPVRIIDI-------------EGVD-----ANMCCGTHVRNLSDLQMIKILGTEKG 230
Query 144 AKGIRRIVAVAGE 156
K ++ ++GE
Sbjct 231 KKNKTNLIFLSGE 243
> dre:445178 aarsd1, im:7152658, zgc:101066; alanyl-tRNA synthetase
domain containing 1; K07050
Length=412
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 52/116 (44%), Gaps = 26/116 (22%)
Query 44 QIEAIELYVQRAIASSLPVNTAVLPLKTAMELPALCAVFGEVYPD----PVRVVSIGPDT 99
++EA+E V I + +PV +L +++ PA+ V PD P+R++ I
Sbjct 149 EMEALETAVNEKIRAHVPVTVNLL----SIDDPAVEKVRSRGLPDDHAGPIRIIDI---- 200
Query 100 KELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESIAKGIRRIVAVAG 155
+G D +C GTH+SN S L+ ++ E K ++ +AG
Sbjct 201 ---------EGVD-----ANMCCGTHVSNLSHLQVIKILGTEKGKKNKTNLIFIAG 242
> mmu:71831 1700007B14Rik, MGC143580, MGC143581; RIKEN cDNA 1700007B14
gene
Length=489
Score = 34.7 bits (78), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 3/64 (4%)
Query 62 VNTAVLPLKTAMELPALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQ---GADKINASV 118
+ ++ LP +A L F Y P+R + P KELKT + ++ +DKI++ V
Sbjct 198 IRSSYLPHGSAQSLSESSVTFKNYYKRPIRKKDLQPIKKELKTEKRSKTIAKSDKIDSKV 257
Query 119 ELCG 122
+ G
Sbjct 258 KRIG 261
> cel:T19A5.5 nhr-219; Nuclear Hormone Receptor family member
(nhr-219)
Length=511
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 23 SLVEERRLRFDFDWDAPLTNQQIEA 47
SLV + ++FDFDW + L+N Q++A
Sbjct 362 SLVNTQTVKFDFDWLSHLSNAQMKA 386
> cel:T14G8.3 hypothetical protein; K09486 hypoxia up-regulated
1
Length=921
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 9/86 (10%)
Query 76 PALCAVFGEVYPDPVRVVSIGPDTKELKTIQETQGADKINAS-VELCGGTHLSNASQLED 134
P ++FGE YP P RV+ + + K + Q ADK S VE+ G + +A + E
Sbjct 452 PIRKSLFGENYPVPNRVMHFSSYSDDFKI--DIQDADKNPLSTVEISG---VKDAIEKE- 505
Query 135 FVVISEESIAKGIRRIVAVAGEAAVS 160
V E S+ KG++ ++ VS
Sbjct 506 --VTDENSVLKGVKTTFSIDLSGIVS 529
> mmu:68592 Syf2, 1110018L13Rik, Cbpin, D4Bwg1551e, Gcipip, mp29,
p29; SYF2 homolog, RNA splicing factor (S. cerevisiae);
K12868 pre-mRNA-splicing factor SYF2
Length=242
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 95 IGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESIAK 145
I PD + + +E G D S L GTH+ ++ +++ V+ E+ I K
Sbjct 141 IKPDMESYERQREKHGEDFFPTSNSLLHGTHVPSSEEIDRMVLDLEKQIEK 191
> ath:AT1G16710 HAC12; HAC12 (histone acetyltransferase of the
CBP family 12); H3/H4 histone acetyltransferase/ histone acetyltransferase/
transcription cofactor; K04498 E1A/CREB-binding
protein [EC:2.3.1.48]
Length=1706
Score = 28.9 bits (63), Expect = 9.1, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 89 PVRVVSIGPDTKELKTIQETQGADKINASVELCGGTH 125
PV + I DTK+ I E++ D A + LC G H
Sbjct 1465 PVEIADIPTDTKDRDEILESEFFDTRQAFLSLCQGNH 1501
> hsa:25949 SYF2, CBPIN, DKFZp564O2082, NTC31, P29, fSAP29; SYF2
homolog, RNA splicing factor (S. cerevisiae); K12868 pre-mRNA-splicing
factor SYF2
Length=243
Score = 28.5 bits (62), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 95 IGPDTKELKTIQETQGADKINASVELCGGTHLSNASQLEDFVVISEESIAK 145
I PD + + ++E G + S L GTH+ + +++ V+ E+ I K
Sbjct 142 IKPDMETYERLREKHGEEFFPTSNSLLHGTHVPSTEEIDRMVIDLEKQIEK 192
Lambda K H
0.313 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3962792044
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40