bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1386_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
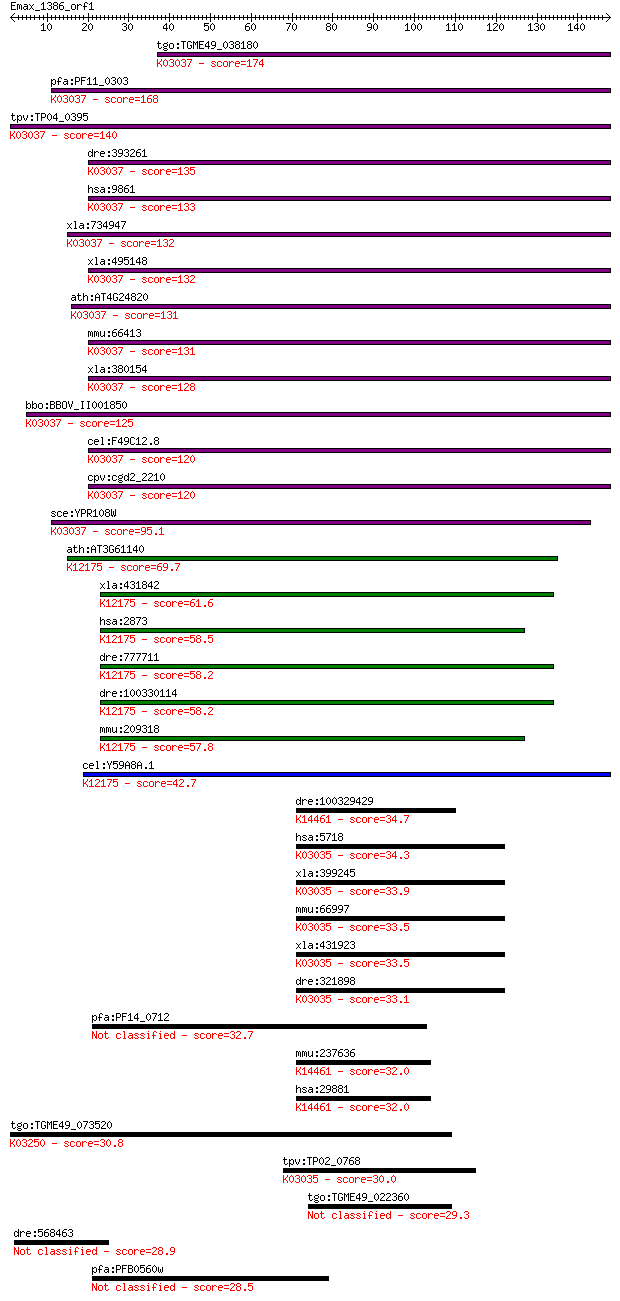
tgo:TGME49_038180 26S proteasome non-ATPase regulatory subunit... 174 1e-43
pfa:PF11_0303 26S proteasome regulatory complex subunit, putat... 168 5e-42
tpv:TP04_0395 26S proteasome regulatory subunit; K03037 26S pr... 140 2e-33
dre:393261 psmd6, MGC56471, zgc:56471; proteasome (prosome, ma... 135 6e-32
hsa:9861 PSMD6, KIAA0107, Rpn7, S10, SGA-113M, p44S10; proteas... 133 3e-31
xla:734947 hypothetical protein MGC131117; K03037 26S proteaso... 132 3e-31
xla:495148 psmd6, p44s10, rpn7, s10; proteasome (prosome, macr... 132 5e-31
ath:AT4G24820 26S proteasome regulatory subunit, putative (RPN... 131 6e-31
mmu:66413 Psmd6, 2400006A19Rik; proteasome (prosome, macropain... 131 6e-31
xla:380154 p44s10; hypothetical protein MGC53209; K03037 26S p... 128 8e-30
bbo:BBOV_II001850 18.m06141; PCI domain containing 26S proteas... 125 6e-29
cel:F49C12.8 rpn-7; proteasome Regulatory Particle, Non-ATPase... 120 1e-27
cpv:cgd2_2210 proteasome regulatory subunit Rpn7/26S proteasom... 120 1e-27
sce:YPR108W RPN7; Essential, non-ATPase regulatory subunit of ... 95.1 6e-20
ath:AT3G61140 FUS6; FUS6 (FUSCA 6); K12175 COP9 signalosome co... 69.7 3e-12
xla:431842 gps1, csn1; G protein pathway suppressor 1; K12175 ... 61.6 8e-10
hsa:2873 GPS1, COPS1, CSN1, MGC71287; G protein pathway suppre... 58.5 7e-09
dre:777711 gps1, im:6909260, im:7145572, zgc:154082; G protein... 58.2 9e-09
dre:100330114 G protein pathway suppressor 1-like; K12175 COP9... 58.2 9e-09
mmu:209318 Gps1, Cops1, Csn1, MGC118012, MGC7191, R75577, Sgn1... 57.8 1e-08
cel:Y59A8A.1 csn-1; COP-9 SigNalosome subunit family member (c... 42.7 4e-04
dre:100329429 NPC1 (Niemann-Pick disease, type C1, gene)-like ... 34.7 0.12
hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, mac... 34.3 0.15
xla:399245 psmd12, MGC80339; proteasome (prosome, macropain) 2... 33.9 0.20
mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (pr... 33.5 0.22
xla:431923 hypothetical protein MGC81129; K03035 26S proteasom... 33.5 0.27
dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteas... 33.1 0.36
pfa:PF14_0712 conserved Plasmodium protein, unknown function 32.7 0.46
mmu:237636 Npc1l1, 9130221N23Rik, Gm243; NPC1-like 1; K14461 N... 32.0 0.62
hsa:29881 NPC1L1, NPC11L1; NPC1 (Niemann-Pick disease, type C1... 32.0 0.64
tgo:TGME49_073520 proteasome PCI domain-containing protein ; K... 30.8 1.8
tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasom... 30.0 2.9
tgo:TGME49_022360 hypothetical protein 29.3 4.5
dre:568463 ryr2b; ryanodine receptor 2b (cardiac) 28.9 6.2
pfa:PFB0560w conserved Plasmodium protein 28.5 7.9
> tgo:TGME49_038180 26S proteasome non-ATPase regulatory subunit,
putative ; K03037 26S proteasome regulatory subunit N7
Length=392
Score = 174 bits (440), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 78/111 (70%), Positives = 97/111 (87%), Gaps = 0/111 (0%)
Query 37 PIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANAFGVSPEFIEAEVSS 96
PIA+RVK D+YL HYLYFIR+IRLRAY Q+L+PYKSVT+ NMA AFGVSP FIEAEVS
Sbjct 280 PIARRVKCDLYLGRHYLYFIRAIRLRAYSQFLEPYKSVTIENMATAFGVSPSFIEAEVSG 339
Query 97 FIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNRIQKLARVL 147
FIASG+ C++DRVN ++ ++RPD+RS SYL I++QGD+LLNRIQKL+RV+
Sbjct 340 FIASGKLSCRIDRVNAVVESNRPDERSRSYLRILKQGDLLLNRIQKLSRVI 390
> pfa:PF11_0303 26S proteasome regulatory complex subunit, putative;
K03037 26S proteasome regulatory subunit N7
Length=393
Score = 168 bits (426), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 76/137 (55%), Positives = 105/137 (76%), Gaps = 0/137 (0%)
Query 11 GAGEDMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDP 70
+ ED+ L++FY YR FM+ + IA RVK D YL HY YFIR+ R+RAY+Q+L+P
Sbjct 255 SSDEDLHTYLHSFYHCDYRTFMEKTIKIAMRVKRDRYLGRHYRYFIRNTRVRAYKQFLEP 314
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPII 130
+K+VTL NMA AFGVS EFIE E+SSFIA+G+ CK+D+VNG I +++P++R+ Y I
Sbjct 315 FKNVTLKNMAFAFGVSEEFIENEISSFIANGKLNCKIDKVNGSIESNQPNERNTMYQNTI 374
Query 131 RQGDVLLNRIQKLARVL 147
++GD+LLNRIQKL+RV+
Sbjct 375 KKGDILLNRIQKLSRVI 391
> tpv:TP04_0395 26S proteasome regulatory subunit; K03037 26S
proteasome regulatory subunit N7
Length=393
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 68/149 (45%), Positives = 104/149 (69%), Gaps = 2/149 (1%)
Query 1 QAVFAPAIAEGA--GEDMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRS 58
+ + + +A+ A G + L++ +Y Y+ +M++LV ++K + D YL H YF+R
Sbjct 243 KVLLSSEVAQVASPGSCLYQLISDYYNCNYKNYMKHLVDVSKLILKDRYLGRHCRYFVRQ 302
Query 59 IRLRAYQQYLDPYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHR 118
RL AY+Q+L PYKSVTL NMA+AF VS EFIE E+ S+I+ R CK+D+VNGII +
Sbjct 303 ARLPAYKQFLRPYKSVTLKNMADAFQVSTEFIEEELVSYISGMRLDCKIDKVNGIIENNV 362
Query 119 PDDRSMSYLPIIRQGDVLLNRIQKLARVL 147
D+R+ +Y+ I+QGD+L+NRIQKL+R++
Sbjct 363 GDERNNNYVKTIKQGDLLINRIQKLSRII 391
> dre:393261 psmd6, MGC56471, zgc:56471; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 6; K03037 26S proteasome
regulatory subunit N7
Length=386
Score = 135 bits (339), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 61/128 (47%), Positives = 89/128 (69%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D +PHY Y++R +R+ AY Q L+ Y+S+TL M
Sbjct 257 LFSLYECRYSVFFQSLARVEQEMKQDWLFAPHYRYYVREMRILAYSQLLESYRSLTLGYM 316
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGVS EFI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 317 AEAFGVSTEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKKGDLLLNR 376
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 377 VQKLSRVI 384
> hsa:9861 PSMD6, KIAA0107, Rpn7, S10, SGA-113M, p44S10; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 6; K03037
26S proteasome regulatory subunit N7
Length=389
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 60/128 (46%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D +PHY Y++R +R+ AY Q L+ Y+S+TL M
Sbjct 260 LFSLYECRYSVFFQSLAVVEQEMKKDWLFAPHYRYYVREMRIHAYSQLLESYRSLTLGYM 319
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGV EFI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 320 AEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKKGDLLLNR 379
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 380 VQKLSRVI 387
> xla:734947 hypothetical protein MGC131117; K03037 26S proteasome
regulatory subunit N7
Length=389
Score = 132 bits (333), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 92/133 (69%), Gaps = 0/133 (0%)
Query 15 DMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSV 74
D+K L + Y +Y F +L + K+++ D + PHY Y++R +R+ Y Q L+ Y+S+
Sbjct 255 DVKQFLFSLYNCQYENFYVHLAGVEKQLRADYLIHPHYRYYVREMRILGYTQLLESYRSL 314
Query 75 TLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGD 134
TL MA AFGV+ ++I+ E++ FIA+GR KVDRV GI+ +RPD+++ Y I+QGD
Sbjct 315 TLQYMAEAFGVTVDYIDQELARFIAAGRLHAKVDRVGGIVETNRPDNKNWQYQATIKQGD 374
Query 135 VLLNRIQKLARVL 147
+LLNRIQKL+RV+
Sbjct 375 LLLNRIQKLSRVI 387
> xla:495148 psmd6, p44s10, rpn7, s10; proteasome (prosome, macropain)
26S subunit, non-ATPase, 6; K03037 26S proteasome regulatory
subunit N7
Length=389
Score = 132 bits (331), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 89/128 (69%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D + HY Y++R +R++AY Q L+ Y+S+TL +M
Sbjct 260 LFSLYECRYSVFFQSLAAVEQELKKDWLFASHYRYYVREMRIQAYGQLLESYRSLTLGHM 319
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGV EFI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 320 AEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKKGDLLLNR 379
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 380 VQKLSRVI 387
> ath:AT4G24820 26S proteasome regulatory subunit, putative (RPN7);
K03037 26S proteasome regulatory subunit N7
Length=387
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 60/132 (45%), Positives = 90/132 (68%), Gaps = 0/132 (0%)
Query 16 MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVT 75
+ LN+ Y +Y+ F +A ++K D YL PH+ +++R +R Y Q+L+ YKSVT
Sbjct 254 LSEFLNSLYECQYKAFFSAFAGMAVQIKYDRYLYPHFRFYMREVRTVVYSQFLESYKSVT 313
Query 76 LINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDV 135
+ MA AFGVS +FI+ E+S FIA+G+ CK+D+V G++ +RPD ++ Y I+QGD
Sbjct 314 VEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRPDAKNALYQATIKQGDF 373
Query 136 LLNRIQKLARVL 147
LLNRIQKL+RV+
Sbjct 374 LLNRIQKLSRVI 385
> mmu:66413 Psmd6, 2400006A19Rik; proteasome (prosome, macropain)
26S subunit, non-ATPase, 6; K03037 26S proteasome regulatory
subunit N7
Length=389
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 88/128 (68%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D +PHY Y++R +R+ AY Q L+ Y+S+TL M
Sbjct 260 LFSLYECRYSVFFQSLAIVEQEMKKDWLFAPHYRYYVREMRIHAYSQLLESYRSLTLGYM 319
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGV +FI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 320 AEAFGVGVDFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKKGDLLLNR 379
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 380 VQKLSRVI 387
> xla:380154 p44s10; hypothetical protein MGC53209; K03037 26S
proteasome regulatory subunit N7
Length=389
Score = 128 bits (321), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 87/128 (67%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
L + Y +Y F Q L + + +K D + HY Y++R +R++ Y Q L+ Y+S+TL M
Sbjct 260 LFSMYECRYAVFFQSLAAVEQELKEDWLFASHYRYYVREMRIQTYGQLLESYRSLTLGYM 319
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A AFGV EFI+ E+S FIA+GR CK+D+VN I+ +RPD ++ Y I++GD+LLNR
Sbjct 320 AEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDSKNWQYQETIKRGDLLLNR 379
Query 140 IQKLARVL 147
+QKL+RV+
Sbjct 380 VQKLSRVI 387
> bbo:BBOV_II001850 18.m06141; PCI domain containing 26S proteasome
non-ATPase regulatory subunit 6 protein; K03037 26S proteasome
regulatory subunit N7
Length=396
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 63/145 (43%), Positives = 96/145 (66%), Gaps = 2/145 (1%)
Query 5 APAIAEGAGED--MKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLR 62
+P +++ A E + LL FY Y+ +M L + D Y+S H Y +R RL
Sbjct 250 SPEVSQVATEGSLLHQLLYDFYHCNYKNYMFNLARTYDLILRDKYMSRHCKYILRQARLP 309
Query 63 AYQQYLDPYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDR 122
AY+Q+L PYKSVT+ NM++AF + P+FIE E+ S+I+ R CK+D VNGII + D+R
Sbjct 310 AYRQFLRPYKSVTIENMSHAFQLPPDFIEDELVSYISGMRLDCKIDLVNGIIENNIMDER 369
Query 123 SMSYLPIIRQGDVLLNRIQKLARVL 147
+ +Y+ I+++GD+LLNRIQKL+R++
Sbjct 370 NTNYIEIVKEGDMLLNRIQKLSRIV 394
> cel:F49C12.8 rpn-7; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-7); K03037 26S proteasome regulatory
subunit N7
Length=410
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 60/129 (46%), Positives = 86/129 (66%), Gaps = 1/129 (0%)
Query 20 LNAFYFAKY-REFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
L ++Y Y R F+Q ++R K D YLSPH+ Y+ R +R RAY+Q+L PYK+V +
Sbjct 280 LESYYDCHYDRFFIQLAALESERFKFDRYLSPHFNYYSRGMRHRAYEQFLTPYKTVRIDM 339
Query 79 MANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
MA FGVS FI+ E+ IA+G+ C++D VNG+I + D ++ Y +I+ GD+LLN
Sbjct 340 MAKDFGVSRAFIDRELHRLIATGQLQCRIDAVNGVIEVNHRDSKNHLYKAVIKDGDILLN 399
Query 139 RIQKLARVL 147
RIQKLARV+
Sbjct 400 RIQKLARVI 408
> cpv:cgd2_2210 proteasome regulatory subunit Rpn7/26S proteasome
subunit 6, PINT domain containing protein ; K03037 26S proteasome
regulatory subunit N7
Length=403
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 90/128 (70%), Gaps = 0/128 (0%)
Query 20 LNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINM 79
+ +FY+ +Y +F + LV I ++ D Y + H+ Y++R +R +AY QYL+PY+SV++++M
Sbjct 274 IESFYYGRYSQFFERLVNITYILQKDYYFNRHHKYYLRIVRSKAYIQYLEPYESVSILSM 333
Query 80 ANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLNR 139
A +FG++ EF+E ++ +FI+S + C +D+V +I R D + Y ++++GD+LLNR
Sbjct 334 AESFGITQEFLEKDIVTFISSSKLSCTIDKVKQVIICSRNDKKINQYNELVQKGDLLLNR 393
Query 140 IQKLARVL 147
+Q+L R++
Sbjct 394 LQRLTRII 401
> sce:YPR108W RPN7; Essential, non-ATPase regulatory subunit of
the 26S proteasome, similar to another S. cerevisiae regulatory
subunit, Rpn5p, as well as to mammalian proteasome subunits;
K03037 26S proteasome regulatory subunit N7
Length=429
Score = 95.1 bits (235), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 81/133 (60%), Gaps = 1/133 (0%)
Query 11 GAGEDMKGLLNAFYFAKYREFMQYLVPIAKRVKTDI-YLSPHYLYFIRSIRLRAYQQYLD 69
A + + L + Y + Y + YL+ V YL+ H +F+R +R + Y Q L+
Sbjct 284 AALQSISSLTISLYASDYASYFPYLLETYANVLIPCKYLNRHADFFVREMRRKVYAQLLE 343
Query 70 PYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPI 129
YK+++L +MA+AFGVS F++ ++ FI + + C +DRVNGI+ +RPD+++ Y +
Sbjct 344 SYKTLSLKSMASAFGVSVAFLDNDLGKFIPNKQLNCVIDRVNGIVETNRPDNKNAQYHLL 403
Query 130 IRQGDVLLNRIQK 142
++QGD LL ++QK
Sbjct 404 VKQGDGLLTKLQK 416
> ath:AT3G61140 FUS6; FUS6 (FUSCA 6); K12175 COP9 signalosome
complex subunit 1
Length=441
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 63/120 (52%), Gaps = 0/120 (0%)
Query 15 DMKGLLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSV 74
D++ L+N FY ++Y ++YL + + DI+L H IR +A QY P+ SV
Sbjct 294 DVRELINDFYSSRYASCLEYLASLKSNLLLDIHLHDHVDTLYDQIRKKALIQYTLPFVSV 353
Query 75 TLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGD 134
L MA+AF S +E E+ + I + ++D N I+ A D R+ ++ +++ G+
Sbjct 354 DLSRMADAFKTSVSGLEKELEALITDNQIQARIDSHNKILYARHADQRNATFQKVLQMGN 413
> xla:431842 gps1, csn1; G protein pathway suppressor 1; K12175
COP9 signalosome complex subunit 1
Length=487
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L I + D+YL+PH IR RA QY PY S + MA A
Sbjct 329 FYESKYASCLKMLDEIKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMYKMATA 388
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQG 133
F + +E E++ I G ++D + I+ A D RS ++ ++ G
Sbjct 389 FNTTVSALEDELTQLILEGLINARIDSHSKILYARDVDQRSTTFEKSLQMG 439
> hsa:2873 GPS1, COPS1, CSN1, MGC71287; G protein pathway suppressor
1; K12175 COP9 signalosome complex subunit 1
Length=491
Score = 58.5 bits (140), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 51/104 (49%), Gaps = 0/104 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L + + D+YL+PH IR RA QY PY S + MA A
Sbjct 333 FYESKYASCLKMLDEMKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMHRMAAA 392
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSY 126
F + +E E++ I G +VD + I+ A D RS ++
Sbjct 393 FNTTVAALEDELTQLILEGLISARVDSHSKILYARDVDQRSTTF 436
> dre:777711 gps1, im:6909260, im:7145572, zgc:154082; G protein
pathway suppressor 1; K12175 COP9 signalosome complex subunit
1
Length=487
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L + + D+YL+PH IR RA QY PY S + MA A
Sbjct 329 FYESKYASCLKMLDEMKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMNKMAVA 388
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQG 133
F + +E E++ I G ++D + I+ A D RS ++ ++ G
Sbjct 389 FNTTVAALEDELTQLILEGLINARIDSHSKILYARDVDQRSTTFEKSLQMG 439
> dre:100330114 G protein pathway suppressor 1-like; K12175 COP9
signalosome complex subunit 1
Length=487
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L + + D+YL+PH IR RA QY PY S + MA A
Sbjct 329 FYESKYASCLKMLDEMKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMNKMAVA 388
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQG 133
F + +E E++ I G ++D + I+ A D RS ++ ++ G
Sbjct 389 FNTTVAALEDELTQLILEGLINARIDSHSKILYARDVDQRSTTFEKSLQMG 439
> mmu:209318 Gps1, Cops1, Csn1, MGC118012, MGC7191, R75577, Sgn1;
G protein pathway suppressor 1; K12175 COP9 signalosome
complex subunit 1
Length=487
Score = 57.8 bits (138), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 51/104 (49%), Gaps = 0/104 (0%)
Query 23 FYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLINMANA 82
FY +KY ++ L + + D+YL+PH IR RA QY PY S + MA A
Sbjct 329 FYESKYASCLKMLDEMKDNLLLDMYLAPHVRTLYTQIRNRALIQYFSPYVSADMHKMAAA 388
Query 83 FGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSY 126
F + +E E++ I G ++D + I+ A D RS ++
Sbjct 389 FNTTVAALEDELTQLILEGLINARIDSHSKILYARDVDQRSTTF 432
> cel:Y59A8A.1 csn-1; COP-9 SigNalosome subunit family member
(csn-1); K12175 COP9 signalosome complex subunit 1
Length=601
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 28/129 (21%), Positives = 59/129 (45%), Gaps = 4/129 (3%)
Query 19 LLNAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
LL ++ +++ + + + R+ D ++S + IR + QYL PY ++ +
Sbjct 398 LLGSYTSSRFGRCFEIMRSVKPRLLLDPFISRNVDELFEKIRQKCVLQYLQPYSTIKMAT 457
Query 79 MANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDDRSMSYLPIIRQGDVLLN 138
MA A G+S ++ + I K+D+ GI+ D + I+++ +V +
Sbjct 458 MAEAVGMSSAELQLSLLELIEQKHVSLKIDQNEGIVRILDERDEN----AILKRVNVTCD 513
Query 139 RIQKLARVL 147
R + A+ L
Sbjct 514 RATQKAKSL 522
> dre:100329429 NPC1 (Niemann-Pick disease, type C1, gene)-like
1-like; K14461 Niemann-Pick C1-like protein 1
Length=1383
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 24/39 (61%), Gaps = 1/39 (2%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDR 109
+ +V+LIN+ A G+S EF+ SF S RP KV+R
Sbjct 1178 FNAVSLINLVTAVGISVEFVSHTTRSFALSVRPT-KVER 1215
> hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome
regulatory subunit N5
Length=456
Score = 34.3 bits (77), Expect = 0.15, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ MA +S + EA +S+ + + KVDR+ GII RP D
Sbjct 370 YTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLAGIINFQRPKD 420
> xla:399245 psmd12, MGC80339; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome regulatory
subunit N5
Length=441
Score = 33.9 bits (76), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ MA +S + E +SS + + KVDR+ GII RP D
Sbjct 355 YTRITMKRMAQLLDLSIDESEEFLSSLVVNKTIYAKVDRLAGIINFQRPKD 405
> mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 12; K03035 26S
proteasome regulatory subunit N5
Length=456
Score = 33.5 bits (75), Expect = 0.22, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ MA +S + EA +S+ + + KVDR+ G+I RP D
Sbjct 370 YTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLAGVINFQRPKD 420
> xla:431923 hypothetical protein MGC81129; K03035 26S proteasome
regulatory subunit N5
Length=441
Score = 33.5 bits (75), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y +T+ MA +S E +SS + + KVDR+ GII RP D
Sbjct 355 YTRITMKRMAQLLDLSVNESEEFLSSLVVNKTIYAKVDRLAGIINFQRPKD 405
> dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 12; K03035
26S proteasome regulatory subunit N5
Length=456
Score = 33.1 bits (74), Expect = 0.36, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGIIAAHRPDD 121
Y S+T+ MA +S + E +S+ + + KVDR+ GII RP D
Sbjct 370 YTSITMGRMAALLDLSVDESEEFLSNLVVNKTIYAKVDRLAGIINFQRPKD 420
> pfa:PF14_0712 conserved Plasmodium protein, unknown function
Length=2977
Score = 32.7 bits (73), Expect = 0.46, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 48/93 (51%), Gaps = 11/93 (11%)
Query 21 NAFYFAKY---REFMQYLVPIAKRV----KTDIYLSPHYLYFIRS--IRLRAYQQYLDPY 71
N FY Y +E +Y + I K++ DI+L H++Y + I+L Y ++ Y
Sbjct 2220 NKFYMYFYNILKEKKEYCLNIKKKLFLDFYIDIFLFNHFIYSNKCSFIKLYLYSDNINIY 2279
Query 72 KSVTLINMANAFGVSPEFIEAEVSS--FIASGR 102
K + +NM N+ + FI +++ S F++S +
Sbjct 2280 KDIISLNMNNSTYNNQIFIFSKIKSLHFVSSHK 2312
> mmu:237636 Npc1l1, 9130221N23Rik, Gm243; NPC1-like 1; K14461
Niemann-Pick C1-like protein 1
Length=1333
Score = 32.0 bits (71), Expect = 0.62, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRP 103
Y +V+LIN+ A G+S EF+ SF S +P
Sbjct 1164 YNAVSLINLVTAVGMSVEFVSHITRSFAVSTKP 1196
> hsa:29881 NPC1L1, NPC11L1; NPC1 (Niemann-Pick disease, type
C1, gene)-like 1; K14461 Niemann-Pick C1-like protein 1
Length=1332
Score = 32.0 bits (71), Expect = 0.64, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 71 YKSVTLINMANAFGVSPEFIEAEVSSFIASGRP 103
Y +V+LIN+ +A G+S EF+ SF S +P
Sbjct 1163 YNAVSLINLVSAVGMSVEFVSHITRSFAISTKP 1195
> tgo:TGME49_073520 proteasome PCI domain-containing protein ;
K03250 translation initiation factor 3 subunit E
Length=601
Score = 30.8 bits (68), Expect = 1.8, Method: Composition-based stats.
Identities = 26/112 (23%), Positives = 46/112 (41%), Gaps = 5/112 (4%)
Query 1 QAVFAPAIAEGA---GEDMKGLLNAFYFA-KYREFMQYLVPIAKRVKTDIYLSPHYLYFI 56
QAV AI +G + LL A + + E ++ I + +D +L P
Sbjct 369 QAVIT-AIGQGKNRYSDPFTSLLEALFLDFNFDEAQHHIARIGEECDSDFFLQPLKGAIN 427
Query 57 RSIRLRAYQQYLDPYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVD 108
RL ++ Y +K + + +A +SPE E + + I R ++D
Sbjct 428 EFARLLIFETYCRIHKCINVDMIAKKVNMSPEAAERWIVNLIRHARLEARID 479
> tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasome
regulatory subunit N5
Length=500
Score = 30.0 bits (66), Expect = 2.9, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 68 LDPYKSVTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVDRVNGII 114
++PY S+TL+ + S +E E+S+ + G K++R+ GI+
Sbjct 399 VNPYYSITLVTHSIHLLPSKSKLENEISNMVEMGIIDSKINRITGIV 445
> tgo:TGME49_022360 hypothetical protein
Length=808
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 74 VTLINMANAFGVSPEFIEAEVSSFIASGRPVCKVD 108
V +I MA A GVSP+ +EA+ R + +D
Sbjct 73 VNVIEMATALGVSPDIVEAKTEEMTRRSRHLMLLD 107
> dre:568463 ryr2b; ryanodine receptor 2b (cardiac)
Length=4882
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 2 AVFAPAIAEGAGEDMKGLLNAFY 24
A FA A+ + AGE +G+LNAFY
Sbjct 506 AHFAEAVGKEAGECWEGVLNAFY 528
> pfa:PFB0560w conserved Plasmodium protein
Length=3990
Score = 28.5 bits (62), Expect = 7.9, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 27/58 (46%), Gaps = 6/58 (10%)
Query 21 NAFYFAKYREFMQYLVPIAKRVKTDIYLSPHYLYFIRSIRLRAYQQYLDPYKSVTLIN 78
N F K RE ++ V I K K Y LY + I L YLDP+ +V LIN
Sbjct 3511 NLFEITKVREILKTKVSILKNNK--YYERIKLLYLLNCIPL----IYLDPHMNVILIN 3562
Lambda K H
0.326 0.141 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40