bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
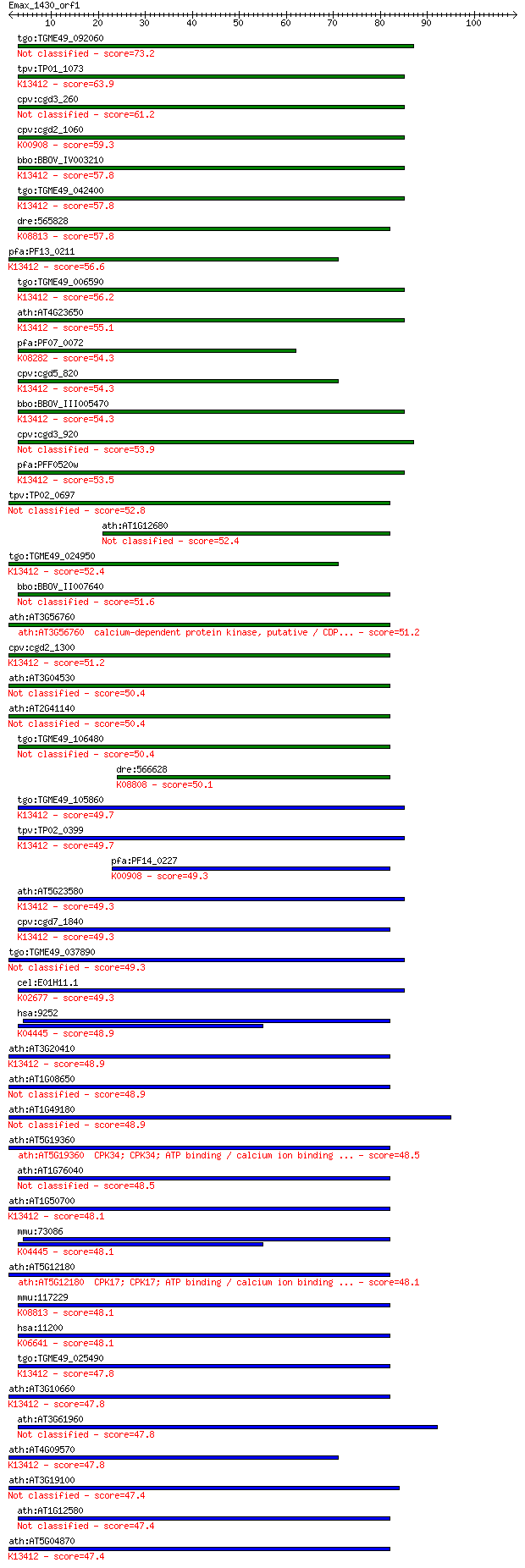
Query= Emax_1430_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_092060 protein kinase, putative (EC:2.7.11.25 2.7.1... 73.2 2e-13
tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium... 63.9 1e-10
cpv:cgd3_260 hypothetical protein 61.2 7e-10
cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with... 59.3 3e-09
bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase ... 57.8 7e-09
tgo:TGME49_042400 calcium-dependent protein kinase, putative (... 57.8 8e-09
dre:565828 stk33, si:ch211-208c6.6, wu:fab03g11; serine/threon... 57.8 8e-09
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 56.6 2e-08
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 56.2 3e-08
ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6... 55.1 6e-08
pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:... 54.3 8e-08
cpv:cgd5_820 calcium/calmodulin dependent protein kinase with ... 54.3 9e-08
bbo:BBOV_III005470 17.m07489; protein kinase domain containing... 54.3 1e-07
cpv:cgd3_920 calmodulin-domain protein kinase 1 53.9
pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7... 53.5 1e-07
tpv:TP02_0697 calmodulin-domain protein kinase 52.8 3e-07
ath:AT1G12680 PEPKR2; PEPKR2 (Phosphoenolpyruvate carboxylase-... 52.4 3e-07
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 52.4 4e-07
bbo:BBOV_II007640 18.m06635; calcium-dependent protein kinase 51.6 5e-07
ath:AT3G56760 calcium-dependent protein kinase, putative / CDP... 51.2 8e-07
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 51.2 8e-07
ath:AT3G04530 PPCK2; PPCK2; kinase/ protein serine/threonine k... 50.4 1e-06
ath:AT2G41140 CRK1; CRK1 (CDPK-RELATED KINASE 1); calcium ion ... 50.4 1e-06
tgo:TGME49_106480 protein kinase domain-containing protein, pu... 50.4 1e-06
dre:566628 protein serine kinase H2-like; K08808 protein serin... 50.1 2e-06
tgo:TGME49_105860 calcium-dependent protein kinase 1, putative... 49.7 2e-06
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 49.7 2e-06
pfa:PF14_0227 calcium/calmodulin-dependent protein kinase, put... 49.3 3e-06
ath:AT5G23580 CDPK9; CDPK9 (CALMODULIN-LIKE DOMAIN PROTEIN KIN... 49.3 3e-06
cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with... 49.3 3e-06
tgo:TGME49_037890 calcium-dependent protein kinase, putative (... 49.3 3e-06
cel:E01H11.1 pkc-2; Protein Kinase C family member (pkc-2); K0... 49.3 3e-06
hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protei... 48.9 4e-06
ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);... 48.9 4e-06
ath:AT1G08650 PPCK1; PPCK1 (PHOSPHOENOLPYRUVATE CARBOXYLASE KI... 48.9 4e-06
ath:AT1G49180 protein kinase family protein 48.9 4e-06
ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding ... 48.5 5e-06
ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding ... 48.5 6e-06
ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding ... 48.1 6e-06
mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC... 48.1 6e-06
ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding ... 48.1 6e-06
mmu:117229 Stk33, 4921505G21Rik, AW061264; serine/threonine ki... 48.1 7e-06
hsa:11200 CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53; CHK2... 48.1 7e-06
tgo:TGME49_025490 calcium-dependent protein kinase, putative (... 47.8 8e-06
ath:AT3G10660 CPK2; CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDP... 47.8 9e-06
ath:AT3G61960 protein kinase family protein 47.8 9e-06
ath:AT4G09570 CPK4; CPK4; calmodulin-dependent protein kinase/... 47.8 1e-05
ath:AT3G19100 calcium-dependent protein kinase, putative / CDP... 47.4 1e-05
ath:AT1G12580 PEPKR1; PEPKR1 (Phosphoenolpyruvate carboxylase-... 47.4 1e-05
ath:AT5G04870 CPK1; CPK1 (CALCIUM DEPENDENT PROTEIN KINASE 1);... 47.4 1e-05
> tgo:TGME49_092060 protein kinase, putative (EC:2.7.11.25 2.7.11.1)
Length=624
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/85 (44%), Positives = 53/85 (62%), Gaps = 2/85 (2%)
Query 3 GTLYYRSPWQILHGVEGDE-RSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSF 61
GTLYY SP+ +L G+ DE +SD+WA GV L++L SG PPF G+ +A + I T ++F
Sbjct 429 GTLYYLSPF-LLKGLPYDEVQSDMWAAGVILYMLISGAPPFEGNSEAEVSSKILTTHIAF 487
Query 62 SGGVWASLSLPCLGFLRALLYNPLL 86
W +S C +R LL+NP L
Sbjct 488 KEPCWRHVSEECKDLIRRLLHNPFL 512
> tpv:TP01_1073 calmodulin-domain protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=504
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 32/83 (38%), Positives = 50/83 (60%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L GV DE+ D+W++GV L+IL SG+PPFNG ++ I + + SF
Sbjct 212 GTAYYIAP-EVLKGVY-DEKCDVWSIGVILYILLSGYPPFNGQSESEIIKKVQLGKYSFE 269
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W ++ ++ +L YNP
Sbjct 270 MSHWQKVAESAKDLIKKMLIYNP 292
> cpv:cgd3_260 hypothetical protein
Length=1180
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 46/83 (55%), Gaps = 2/83 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GTLYY +P +IL G D++ D+W++GV IL G PPF G+ I + VSFS
Sbjct 388 GTLYYIAP-EILLGKGYDDKCDIWSIGVMAHILLCGVPPFAGNTDTEIIEKVRRGSVSFS 446
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
+W+ +S FL LL NP
Sbjct 447 ESIWSQVSTQAKDFLLQLLNKNP 469
> cpv:cgd2_1060 calcium/calmodulin dependent protein kinase with
a kinas domain and 4 calmodulin-like EF hands ; K00908 Ca2+/calmodulin-dependent
protein kinase [EC:2.7.11.17]
Length=718
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 49/83 (59%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L G + D++ D+W+LGV L+IL G+PPF+G + + I + +F
Sbjct 361 GTPYYVAP-EVLQG-KYDKQCDMWSLGVILYILLCGYPPFHGSNDSIILHKVQKGVYAFK 418
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W +S + +R LL YNP
Sbjct 419 EEDWKHVSFLAIDLIRKLLTYNP 441
> bbo:BBOV_IV003210 21.m02835; calcium-dependent protein kinase
4 (EC:2.7.11.1); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=517
Score = 57.8 bits (138), Expect = 7e-09, Method: Composition-based stats.
Identities = 32/83 (38%), Positives = 45/83 (54%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P +L GV D + D+W+ GV L+IL G PPFNG +++ I + + SF
Sbjct 226 GTAYYIAP-DVLKGVY-DSKCDIWSAGVILYILLCGFPPFNGANESEIIKKVQAGKYSFD 283
Query 63 GGVWASLSLPCLGFL-RALLYNP 84
W +S + R L YNP
Sbjct 284 MPQWRKVSESAKDLIARMLTYNP 306
> tgo:TGME49_042400 calcium-dependent protein kinase, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=604
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 49/83 (59%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P Q+L G + + D+W+ GV ++IL G+PPF+GD+ A I + SF+
Sbjct 288 GTPYYVAP-QVLQG-KYTYKCDIWSAGVIMYILLCGYPPFHGDNDAEILAKVKSGKFSFN 345
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W ++S+ +R LL Y+P
Sbjct 346 EQDWKNVSVEAKDLIRKLLTYDP 368
> dre:565828 stk33, si:ch211-208c6.6, wu:fab03g11; serine/threonine
kinase 33 (EC:2.7.11.1); K08813 serine/threonine kinase
33 [EC:2.7.11.1]
Length=500
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT Y +P ++++G + ++ DLW++GV +++L G PPF K + I ++FS
Sbjct 217 GTPIYMAP-EVVNGHQYSQQCDLWSIGVIMYMLLCGEPPFGSSSKERLSEMIMKGELTFS 275
Query 63 GGVWASLSLPCLGFLRALL 81
G VW ++S +R LL
Sbjct 276 GPVWNTISDAAKNVIRCLL 294
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 41/70 (58%), Gaps = 2/70 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V GT YY +P +IL G + D+R D+W+ GV ++IL G+PPFNG + I + +
Sbjct 282 VVGTPYYIAP-EILRG-KYDKRCDIWSSGVIMYILLCGYPPFNGKNNDEILKKVEKGEFV 339
Query 61 FSGGVWASLS 70
F WA +S
Sbjct 340 FDSNYWARVS 349
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 46/83 (55%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P Q+L G + D R D W+LGV L+IL G+PPF G+ A + + SFS
Sbjct 384 GTPYYVAP-QVLQG-KYDFRCDAWSLGVILYILLCGYPPFYGETDAEVLAKVKTGVFSFS 441
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
G W +S +R L+ NP
Sbjct 442 GPEWKRVSEEARELIRHLININP 464
> ath:AT4G23650 CDPK6; CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE
6); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=529
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 32/83 (38%), Positives = 48/83 (57%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
G+ YY +P ++L G E +D+W+ GV L+IL SG PPF G+++ I AI + FS
Sbjct 241 GSAYYVAP-EVLKRNYGPE-ADIWSAGVILYILLSGVPPFWGENETGIFDAILQGQLDFS 298
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W +LS +R +L Y+P
Sbjct 299 ADPWPALSDGAKDLVRKMLKYDP 321
> pfa:PF07_0072 PfCDPK4; calcium-dependent protein kinase 4 (EC:2.7.1.11);
K08282 non-specific serine/threonine protein kinase
[EC:2.7.11.1]
Length=528
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 2/59 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSF 61
GT YY +P +LHG DE+ D+W+ GV L+IL SG PPFNG ++ I + + +F
Sbjct 233 GTAYYIAP-DVLHGTY-DEKCDIWSCGVILYILLSGCPPFNGSNEYDILKKVEAGKYTF 289
> cpv:cgd5_820 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=523
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 26/68 (38%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L + DE+ D+W++GV LFIL +G+PPF G I R + +F
Sbjct 223 GTAYYIAP-EVLRK-KYDEKCDVWSIGVILFILLAGYPPFGGQTDQEILRKVEKGKYTFD 280
Query 63 GGVWASLS 70
W ++S
Sbjct 281 SPEWKNVS 288
> bbo:BBOV_III005470 17.m07489; protein kinase domain containing
protein; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=755
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 48/83 (57%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L G D+ D+W+ GV +FI+ G+PPF+G+D A+I + ++F
Sbjct 459 GTPYYVAP-EVLIG-SYDKSCDMWSAGVIMFIMLVGYPPFHGNDNATILNNVKRGAINFV 516
Query 63 GGVWASLSLPCLGFL-RALLYNP 84
W +S + + + L Y+P
Sbjct 517 PHHWKHISKSAVDLITKCLSYDP 539
> cpv:cgd3_920 calmodulin-domain protein kinase 1
Length=538
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 30/85 (35%), Positives = 48/85 (56%), Gaps = 3/85 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L G DE+ D+W+ GV L+IL SG PPF G ++ I + + +F
Sbjct 237 GTAYYIAP-EVLRGTY-DEKCDVWSAGVILYILLSGTPPFYGKNEYDILKRVETGKYAFD 294
Query 63 GGVWASLSLPCLGFLRALL-YNPLL 86
W ++S +R +L ++P L
Sbjct 295 LPQWRTISDDAKDLIRKMLTFHPSL 319
> pfa:PFF0520w PfCDPK2; calcium-dependent protein kinase (EC:2.7.11.1);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=509
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 44/83 (53%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P Q+L G + D++ D+W+ GV ++ L G+PPF GD + + + F
Sbjct 231 GTPYYVAP-QVLDG-KYDKKCDIWSSGVIMYTLLCGYPPFYGDTDNEVLKKVKKGEFCFY 288
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W S+S + LL YNP
Sbjct 289 ENDWGSISSDAKNLITKLLTYNP 311
> tpv:TP02_0697 calmodulin-domain protein kinase
Length=579
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ GT+YY P +++ G +E+ D+W+ GV ++++ SG PPFN I I +
Sbjct 296 MHGTVYYVDP-EVIDGCY-NEKCDIWSTGVIVYMMLSGSPPFNSGSDKEILWKIKKGNLK 353
Query 61 FSGGVWASLSLPCLGFLRALL 81
F G W S+S F+R LL
Sbjct 354 FEGVRWNSVSELAKEFIRFLL 374
> ath:AT1G12680 PEPKR2; PEPKR2 (Phosphoenolpyruvate carboxylase-related
kinase 2); ATP binding / kinase/ protein kinase/ protein
serine/threonine kinase
Length=470
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 35/61 (57%), Gaps = 0/61 (0%)
Query 21 ERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFSGGVWASLSLPCLGFLRAL 80
E+ D+W+ GV L+ L SG PF GD +I AI + + F+ GVW S+S P L +
Sbjct 276 EKVDVWSAGVLLYALLSGVLPFKGDSLDAIFEAIKNVKLDFNTGVWESVSKPARDLLARM 335
Query 81 L 81
L
Sbjct 336 L 336
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V GT YY +P ++L G G E D+W+ GV L+IL G+PPF+G D I + + S
Sbjct 370 VVGTPYYVAP-EVLFGKYGSE-CDMWSAGVILYILLCGYPPFHGKDNQEILKKVQVGEYS 427
Query 61 FSGGVWASLS 70
F W +S
Sbjct 428 FDPRHWRRVS 437
> bbo:BBOV_II007640 18.m06635; calcium-dependent protein kinase
Length=580
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT+YY P +++ G E+ D+W+ GV +++L SG PPFNG+ I I + F
Sbjct 304 GTVYYVDP-EVIDG-SYSEKCDVWSTGVIVYMLLSGSPPFNGEGDKEILWKIKKGQLRFE 361
Query 63 GGVWASLSLPCLGFLRALL 81
G W ++ F+ LL
Sbjct 362 GVRWVKITDEAKDFITYLL 380
> ath:AT3G56760 calcium-dependent protein kinase, putative / CDPK,
putative; K00924 [EC:2.7.1.-]
Length=577
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++LH G E +D+W++GV +IL G PF ++ I RA+ +
Sbjct 289 IVGSAYYVAP-EVLHRTYGTE-ADMWSIGVIAYILLCGSRPFWARSESGIFRAVLKAEPN 346
Query 61 FSGGVWASLSLPCLGFLRALL 81
F W SLS + F++ LL
Sbjct 347 FEEAPWPSLSPDAVDFVKRLL 367
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 29/81 (35%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V GT YY +P ++L+G + DLW+ GV L+IL G+PPF+G D I R + S
Sbjct 342 VVGTPYYVAP-EVLYG-SYSKLCDLWSAGVILYILLCGYPPFHGKDNVEILRKVKIGQYS 399
Query 61 FSGGVWASLSLPCLGFLRALL 81
W +S ++ LL
Sbjct 400 LEHNSWKYVSDSAKDLIKRLL 420
> ath:AT3G04530 PPCK2; PPCK2; kinase/ protein serine/threonine
kinase
Length=278
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V GT YY +P +++ G + DE+ D+W+ GV ++ + +G PPFNG+ I +I +
Sbjct 171 VVGTPYYVAP-EVVMGRKYDEKVDIWSAGVVIYTMLAGEPPFNGETAEDIFESILRGNLR 229
Query 61 FSGGVWASLSLPCLGFLRALL 81
F + S+S LR ++
Sbjct 230 FPPKKFGSVSSEAKDLLRKMI 250
> ath:AT2G41140 CRK1; CRK1 (CDPK-RELATED KINASE 1); calcium ion
binding / calcium-dependent protein serine/threonine phosphatase/
calmodulin-dependent protein kinase/ kinase
Length=576
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 45/81 (55%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++LH G E +D+W++GV +IL G PF ++ I RA+ +
Sbjct 288 IVGSAYYVAP-EVLHRTYGTE-ADMWSIGVIAYILLCGSRPFWARTESGIFRAVLKAEPN 345
Query 61 FSGGVWASLSLPCLGFLRALL 81
F W SLS + F++ LL
Sbjct 346 FEEAPWPSLSPEAVDFVKRLL 366
> tgo:TGME49_106480 protein kinase domain-containing protein,
putative (EC:2.7.11.1 2.7.11.17)
Length=1388
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 0/79 (0%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT++Y +P ++ SD W+LGV LF + + PPFNG A + R IT +
Sbjct 1235 GTVHYLAPEVLMGNYYDGFASDAWSLGVVLFTMLAAAPPFNGPTHAELTRQITRGEYHMT 1294
Query 63 GGVWASLSLPCLGFLRALL 81
G W +S +R LL
Sbjct 1295 GRAWWRVSSEAKDLIRRLL 1313
> dre:566628 protein serine kinase H2-like; K08808 protein serine
kinase H [EC:2.7.11.1]
Length=421
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 24 DLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFSGGVWASLSLPCLGFLRALL 81
D+WALG+ ++L SG PF+ +A + RAI SF G W S+S GF+ LL
Sbjct 277 DMWALGIITYVLLSGSLPFDHSSRAKLFRAILKGSYSFHGEPWRSVSSLGKGFINGLL 334
> tgo:TGME49_105860 calcium-dependent protein kinase 1, putative
(EC:2.7.11.17); K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=537
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L + DE+ D+W+ GV L+IL G+PPF G I + + SF
Sbjct 238 GTAYYIAP-EVLRK-KYDEKCDVWSCGVILYILLCGYPPFGGQTDQEILKRVEKGKFSFD 295
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W +S ++ +L Y P
Sbjct 296 PPDWTQVSDEAKQLVKLMLTYEP 318
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 44/83 (53%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY +P ++L G D+ D+W+ GV LFIL G+PPF+G + I + + F
Sbjct 559 GTPYYVAP-EVLLG-NYDKACDIWSAGVILFILLVGYPPFHGSNNTEILNNVKRGTLKFV 616
Query 63 GGVWASLSLPCLGFL-RALLYNP 84
W+ +S + + R L Y P
Sbjct 617 EKHWSHVSKSAIDLIKRCLHYIP 639
> pfa:PF14_0227 calcium/calmodulin-dependent protein kinase, putative;
K00908 Ca2+/calmodulin-dependent protein kinase [EC:2.7.11.17]
Length=284
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 23 SDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFSGGVWASLSLPCLGFLRALL 81
SD+WALGV +F + +G PF G + + I + +++ G ++SLS+ + FL+ LL
Sbjct 202 SDIWALGVMVFFMLTGKYPFEGKNTPKVVDEILNKNINWKGKEFSSLSIEAVDFLKRLL 260
> ath:AT5G23580 CDPK9; CDPK9 (CALMODULIN-LIKE DOMAIN PROTEIN KINASE
9); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase
[EC:2.7.11.1]
Length=490
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
G+ YY +P ++LH G E D+W+ GV L+IL G PPF + + I R I + F
Sbjct 185 GSAYYVAP-EVLHKHYGPE-CDVWSAGVILYILLCGFPPFWAESEIGIFRKILQGKLEFE 242
Query 63 GGVWASLSLPCLGFLRALL-YNP 84
W S+S ++ +L NP
Sbjct 243 INPWPSISESAKDLIKKMLESNP 265
> cpv:cgd7_1840 calcium/calmodulin-dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=676
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 42/79 (53%), Gaps = 2/79 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY SP Q+L G+ G E D W+ GV +++L G+PPF+ + + I + +F
Sbjct 365 GTPYYVSP-QVLEGLYGPE-CDEWSAGVMMYVLLCGYPPFSAPTDSEVMLKIREGTFTFP 422
Query 63 GGVWASLSLPCLGFLRALL 81
W ++S +R LL
Sbjct 423 EKDWLNVSPQAESLIRRLL 441
> tgo:TGME49_037890 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=1171
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 47/85 (55%), Gaps = 3/85 (3%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ GT+YY +P +++ G + +E+ DLW++GV +++L SG PPF G I I +
Sbjct 819 MHGTVYYVAP-EVMDG-KYNEKCDLWSIGVIVYMLLSGSPPFTGHGDQEILIKIRRCKYN 876
Query 61 FSGGVWASLSLPCLGFLRALL-YNP 84
G W +S F+ +LL NP
Sbjct 877 MDGPRWRGISEQAKHFIASLLRRNP 901
> cel:E01H11.1 pkc-2; Protein Kinase C family member (pkc-2);
K02677 classical protein kinase C [EC:2.7.11.13]
Length=725
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 33/83 (39%), Positives = 46/83 (55%), Gaps = 6/83 (7%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT Y +P IL+ G + D WA GV LF + +G PPF+G+D+ + AIT+ VS+
Sbjct 554 GTPDYIAPEIILYQPYG-KSVDWWAYGVLLFEMLAGQPPFDGEDEDELFTAITEHNVSYP 612
Query 63 GGVWASLSLPCLGFLRALLY-NP 84
SLS + +ALL NP
Sbjct 613 ----KSLSKEAVSLCKALLIKNP 631
> hsa:9252 RPS6KA5, MGC1911, MSK1, MSPK1, RLPK; ribosomal protein
S6 kinase, 90kDa, polypeptide 5 (EC:2.7.11.1); K04445 mitogen-,stress
activated protein kinases [EC:2.7.11.1]
Length=802
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 43/85 (50%), Gaps = 8/85 (9%)
Query 4 TLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKA-------SIDRAITD 56
TL+Y +P ++L+ DE DLW+LGV L+ + SG PF D++ I + I
Sbjct 585 TLHYAAP-ELLNQNGYDESCDLWSLGVILYTMLSGQVPFQSHDRSLTCTSAVEIMKKIKK 643
Query 57 TPVSFSGGVWASLSLPCLGFLRALL 81
SF G W ++S ++ LL
Sbjct 644 GDFSFEGEAWKNVSQEAKDLIQGLL 668
Score = 34.7 bits (78), Expect = 0.075, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query 3 GTLYYRSPWQILHGVEGDERS-DLWALGVALFILFSGHPPFNGD----DKASIDRAI 54
GT+ Y +P + G G +++ D W+LGV ++ L +G PF D +A I R I
Sbjct 215 GTIEYMAPDIVRGGDSGHDKAVDWWSLGVLMYELLTGASPFTVDGEKNSQAEISRRI 271
> ath:AT3G20410 CPK9; CPK9 (calmodulin-domain protein kinase 9);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=541
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++L G E D+W+ G+ L+IL SG PPF + + I AI + +
Sbjct 252 IVGSAYYVAP-EVLRRRYGKE-VDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGHID 309
Query 61 FSGGVWASLSLPCLGFLRALL 81
F W S+S +R +L
Sbjct 310 FESQPWPSISSSAKDLVRRML 330
> ath:AT1G08650 PPCK1; PPCK1 (PHOSPHOENOLPYRUVATE CARBOXYLASE
KINASE); kinase/ protein serine/threonine kinase
Length=284
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 1/81 (1%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V GT YY +P ++L G E+ DLW+ GV L+ + +G PPF G+ I A+ +
Sbjct 175 VVGTPYYVAP-EVLMGYSYGEKVDLWSAGVVLYTMLAGTPPFYGETAEEIFEAVLRGNLR 233
Query 61 FSGGVWASLSLPCLGFLRALL 81
F ++ +S FLR L+
Sbjct 234 FPTKIFRGVSSMAKDFLRKLI 254
> ath:AT1G49180 protein kinase family protein
Length=408
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/96 (30%), Positives = 50/96 (52%), Gaps = 3/96 (3%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAI-TDTPV 59
V G+ +Y +P ++L +E++D+W++G LF L G+PPF G++ + R I + T +
Sbjct 166 VCGSPFYMAP-EVLQFQRYNEKADMWSVGAILFELLHGYPPFRGNNNVQVLRNIKSSTAL 224
Query 60 SFSGGVWASLSLPCLGFLRALL-YNPLLPLHLRGPP 94
FS + + C+ LL NP L + P
Sbjct 225 PFSRLILQQMHPDCIDVCSRLLSINPAATLGIEDFP 260
> ath:AT5G19360 CPK34; CPK34; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=523
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++L G E +D+W++GV L+IL G PPF + + I AI V
Sbjct 229 IVGSAYYIAP-EVLRRKYGPE-ADIWSIGVMLYILLCGVPPFWAESENGIFNAILSGQVD 286
Query 61 FSGGVWASLSLPCLGFLRALL 81
FS W +S +R +L
Sbjct 287 FSSDPWPVISPQAKDLVRKML 307
> ath:AT1G76040 CPK29; CPK29; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase
Length=323
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 44/79 (55%), Gaps = 2/79 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
G+ YY +P ++LH G E D+W+ GV L+IL SG PPF G+ + +I AI + +
Sbjct 37 GSAYYVAP-EVLHRNYGKE-IDVWSAGVMLYILLSGVPPFWGETEKTIFEAILEGKLDLE 94
Query 63 GGVWASLSLPCLGFLRALL 81
W ++S +R +L
Sbjct 95 TSPWPTISESAKDLIRKML 113
> ath:AT1G50700 CPK33; CPK33; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=521
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++L G E D+W+ G+ L+IL SG PPF + + I AI + +
Sbjct 234 IVGSAYYVAP-EVLKRRYGKE-IDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGEID 291
Query 61 FSGGVWASLSLPCLGFLRALL 81
F W S+S +R +L
Sbjct 292 FESQPWPSISNSAKDLVRRML 312
> mmu:73086 Rps6ka5, 3110005L17Rik, 6330404E13Rik, AI854034, MGC28385,
MSK1, MSPK1, RLPK; ribosomal protein S6 kinase, polypeptide
5 (EC:2.7.11.1); K04445 mitogen-,stress activated protein
kinases [EC:2.7.11.1]
Length=863
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 41/85 (48%), Gaps = 8/85 (9%)
Query 4 TLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKA-------SIDRAITD 56
TL+Y +P + H DE DLW+LGV L+ + SG PF D++ I + I
Sbjct 649 TLHYAAPELLTHNGY-DESCDLWSLGVILYTMLSGQVPFQSHDRSLTCTSAVEIMKKIKK 707
Query 57 TPVSFSGGVWASLSLPCLGFLRALL 81
SF G W ++S ++ LL
Sbjct 708 GDFSFEGEAWKNVSQEAKDLIQGLL 732
Score = 34.3 bits (77), Expect = 0.088, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query 3 GTLYYRSPWQILHGVEGDERS-DLWALGVALFILFSGHPPFNGD----DKASIDRAI 54
GT+ Y +P + G G +++ D W+LGV ++ L +G PF D +A I R I
Sbjct 214 GTIEYMAPDIVRGGDSGHDKAVDWWSLGVLMYELLTGASPFTVDGEKNSQAEISRRI 270
> ath:AT5G12180 CPK17; CPK17; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K00924 [EC:2.7.1.-]
Length=528
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++L G E +D+W++GV L+IL G PPF + + I AI V
Sbjct 234 IVGSAYYIAP-EVLKRKYGPE-ADIWSIGVMLYILLCGVPPFWAESENGIFNAILRGHVD 291
Query 61 FSGGVWASLSLPCLGFLRALL 81
FS W S+S ++ +L
Sbjct 292 FSSDPWPSISPQAKDLVKKML 312
> mmu:117229 Stk33, 4921505G21Rik, AW061264; serine/threonine
kinase 33 (EC:2.7.11.1); K08813 serine/threonine kinase 33 [EC:2.7.11.1]
Length=491
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT Y +P ++++ + ++ D+W++GV +FIL G PPF + + + I + F
Sbjct 281 GTPIYMAP-EVINAHDYSQQCDIWSIGVIMFILLCGEPPFLANSEEKLYELIKKGELRFE 339
Query 63 GGVWASLSLPCLGFLRALL 81
VW S+S L+ L+
Sbjct 340 NPVWESVSDSAKNTLKQLM 358
> hsa:11200 CHEK2, CDS1, CHK2, HuCds1, LFS2, PP1425, RAD53; CHK2
checkpoint homolog (S. pombe) (EC:2.7.11.1); K06641 serine/threonine-protein
kinase Chk2 [EC:2.7.11.1]
Length=586
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 44/82 (53%), Gaps = 3/82 (3%)
Query 3 GTLYYRSPWQILH-GVEGDERS-DLWALGVALFILFSGHPPFNGD-DKASIDRAITDTPV 59
GT Y +P ++ G G R+ D W+LGV LFI SG+PPF+ + S+ IT
Sbjct 429 GTPTYLAPEVLVSVGTAGYNRAVDCWSLGVILFICLSGYPPFSEHRTQVSLKDQITSGKY 488
Query 60 SFSGGVWASLSLPCLGFLRALL 81
+F VWA +S L ++ LL
Sbjct 489 NFIPEVWAEVSEKALDLVKKLL 510
> tgo:TGME49_025490 calcium-dependent protein kinase, putative
(EC:1.6.3.1 2.7.11.17 3.2.1.60); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=711
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 41/79 (51%), Gaps = 2/79 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
GT YY SP Q+L G G E D+W+ GV ++IL G+PPFN +I + +F
Sbjct 399 GTPYYVSP-QVLEGRYGPE-CDVWSAGVMMYILLCGYPPFNAPSDRAIMNKVRAGHYTFP 456
Query 63 GGVWASLSLPCLGFLRALL 81
W+ +SL + LL
Sbjct 457 DSEWSRVSLQAKDLISRLL 475
> ath:AT3G10660 CPK2; CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDPK
ISOFORM 2); calmodulin-dependent protein kinase/ kinase/
protein serine/threonine kinase; K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=646
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V G+ YY +P ++L G E SD+W+ GV ++IL SG PPF + + I + +
Sbjct 347 VVGSPYYVAP-EVLRKRYGPE-SDVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLD 404
Query 61 FSGGVWASLSLPCLGFLRALL 81
FS W S+S +R +L
Sbjct 405 FSSDPWPSISESAKDLVRKML 425
> ath:AT3G61960 protein kinase family protein
Length=584
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASI-DRAITDTPVSF 61
G+ Y +P +I+ + D ++DLW+ G LF L +G PPF+G++ + + DT + F
Sbjct 171 GSPLYMAP-EIIRNQKYDAKADLWSAGAILFQLVTGKPPFDGNNHIQLFHNIVRDTELKF 229
Query 62 SGGVWASLSLPCLGFLRALL-YNPLLPLHLR 91
+ C+ R+LL NP+ L R
Sbjct 230 PEDTRNEIHPDCVDLCRSLLRRNPIERLTFR 260
> ath:AT4G09570 CPK4; CPK4; calmodulin-dependent protein kinase/
kinase/ protein kinase; K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=501
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V G+ YY +P ++L G E D+W+ GV L+IL SG PPF + ++ I R I +
Sbjct 186 VVGSPYYVAP-EVLKKCYGPE-IDVWSAGVILYILLSGVPPFWAETESGIFRQILQGKID 243
Query 61 FSGGVWASLS 70
F W ++S
Sbjct 244 FKSDPWPTIS 253
> ath:AT3G19100 calcium-dependent protein kinase, putative / CDPK,
putative
Length=599
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
+ G+ YY +P ++LH E +D+W++GV +IL G PF ++ I RA+ S
Sbjct 309 IVGSAYYVAP-EVLHRSYTTE-ADVWSIGVIAYILLCGSRPFWARTESGIFRAVLKADPS 366
Query 61 FSGGVWASLSLPCLGFLRALLYN 83
F W SLS F++ LLY
Sbjct 367 FDEPPWPSLSFEAKDFVKRLLYK 389
> ath:AT1G12580 PEPKR1; PEPKR1 (Phosphoenolpyruvate carboxylase-related
kinase 1); ATP binding / kinase/ protein kinase/ protein
serine/threonine kinase
Length=522
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Query 3 GTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVSFS 62
G+ +Y +P + G ++ +D+W+ GV L+IL SG PPF G K+ I A+ + FS
Sbjct 207 GSPFYIAPEVLAGGY--NQAADVWSAGVILYILLSGAPPFWGKTKSKIFDAVRAADLRFS 264
Query 63 GGVWASLSLPCLGFLRALL 81
W +++ +R +L
Sbjct 265 AEPWDNITSYAKDLIRGML 283
> ath:AT5G04870 CPK1; CPK1 (CALCIUM DEPENDENT PROTEIN KINASE 1);
calmodulin-dependent protein kinase/ kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=610
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 2/81 (2%)
Query 1 VEGTLYYRSPWQILHGVEGDERSDLWALGVALFILFSGHPPFNGDDKASIDRAITDTPVS 60
V G+ YY +P ++L G E +D+W+ GV ++IL SG PPF + + I + +
Sbjct 311 VVGSPYYVAP-EVLRKRYGPE-ADVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLD 368
Query 61 FSGGVWASLSLPCLGFLRALL 81
FS W S+S +R +L
Sbjct 369 FSSDPWPSISESAKDLVRKML 389
Lambda K H
0.321 0.144 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40