bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1488_orf2
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
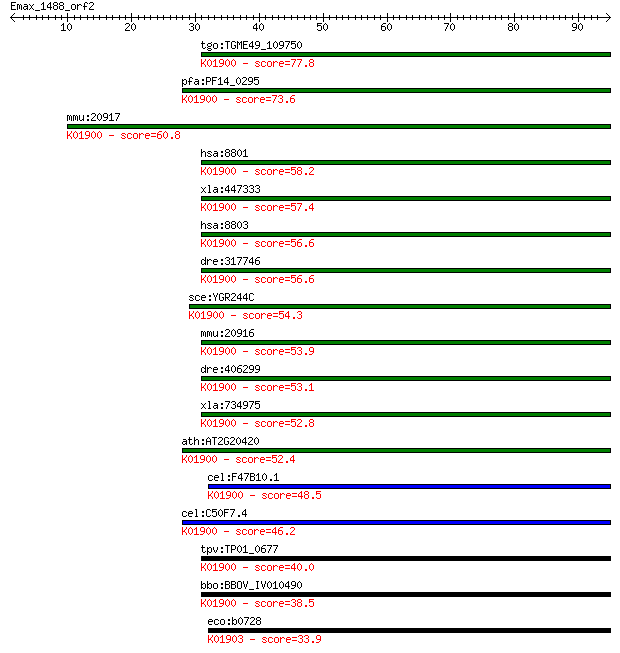
tgo:TGME49_109750 succinyl-CoA ligase, putative (EC:6.2.1.5); ... 77.8 7e-15
pfa:PF14_0295 ATP-specific succinyl-CoA synthetase beta subuni... 73.6 1e-13
mmu:20917 Suclg2, AF171077, AW556404, D6Wsu120e, MGC91183; suc... 60.8 1e-09
hsa:8801 SUCLG2, GBETA; succinate-CoA ligase, GDP-forming, bet... 58.2 7e-09
xla:447333 sucla2, MGC82958; succinate-CoA ligase, ADP-forming... 57.4 1e-08
hsa:8803 SUCLA2, A-BETA, MTDPS5, SCS-betaA; succinate-CoA liga... 56.6 2e-08
dre:317746 suclg2, cb625, wu:fd44e06; succinate-CoA ligase, GD... 56.6 2e-08
sce:YGR244C LSC2; Beta subunit of succinyl-CoA ligase, which i... 54.3 9e-08
mmu:20916 Sucla2, 4930547K18Rik; succinate-Coenzyme A ligase, ... 53.9 1e-07
dre:406299 sucla2, wu:fb18c11, wu:fj38c11, wu:fj90d07, zgc:733... 53.1 2e-07
xla:734975 suclg2, MGC132166, gbeta; succinate-CoA ligase, GDP... 52.8 3e-07
ath:AT2G20420 succinyl-CoA ligase (GDP-forming) beta-chain, mi... 52.4 4e-07
cel:F47B10.1 hypothetical protein; K01900 succinyl-CoA synthet... 48.5 5e-06
cel:C50F7.4 hypothetical protein; K01900 succinyl-CoA syntheta... 46.2 2e-05
tpv:TP01_0677 ATP-specific succinyl-CoA synthetase beta subuni... 40.0 0.002
bbo:BBOV_IV010490 23.m06275; succinly CoA-ligase beta subunit ... 38.5 0.006
eco:b0728 sucC, ECK0716, JW0717; succinyl-CoA synthetase, beta... 33.9 0.13
> tgo:TGME49_109750 succinyl-CoA ligase, putative (EC:6.2.1.5);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=498
Score = 77.8 bits (190), Expect = 7e-15, Method: Composition-based stats.
Identities = 37/64 (57%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
VVKAQ+LAGGR LGFF+ENGY+GGV C +P+ V VA M+ KTL+TKQT K+G+
Sbjct 123 FVVKAQVLAGGRGLGFFRENGYQGGVQVCESPREVGIVAEKMLGKTLVTKQTGKEGKLCN 182
Query 91 RLLI 94
++L+
Sbjct 183 KVLV 186
> pfa:PF14_0295 ATP-specific succinyl-CoA synthetase beta subunit,
putative; K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=462
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 34/67 (50%), Positives = 48/67 (71%), Gaps = 0/67 (0%)
Query 28 ENALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGR 87
+N LV+KAQ+L+GGR +G+FKEN ++GGVH C N V +A M+N TLITKQ+ +G+
Sbjct 93 DNDLVIKAQVLSGGRGVGYFKENNFEGGVHVCRNSMEVKEIATKMLNNTLITKQSGPEGK 152
Query 88 KVRRLLI 94
K + I
Sbjct 153 KCNTVFI 159
> mmu:20917 Suclg2, AF171077, AW556404, D6Wsu120e, MGC91183; succinate-Coenzyme
A ligase, GDP-forming, beta subunit (EC:6.2.1.4);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=433
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 53/85 (62%), Gaps = 3/85 (3%)
Query 10 VQHQGQQQGQQQQRQQQQENALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVA 69
V + ++ + +R +E +V+KAQILAGGR G F +G KGGVH +PK+V +A
Sbjct 62 VANTAKEALEAAKRLNAKE--IVLKAQILAGGRGKGVFN-SGLKGGVHLTKDPKVVGELA 118
Query 70 NNMINKTLITKQTTKKGRKVRRLLI 94
MI L TKQT K+G KV ++++
Sbjct 119 QQMIGYNLATKQTPKEGVKVNKVMV 143
> hsa:8801 SUCLG2, GBETA; succinate-CoA ligase, GDP-forming, beta
subunit (EC:6.2.1.4); K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=440
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 32/64 (50%), Positives = 43/64 (67%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQILAGGR G F +G KGGVH +P +V +A MI L TKQT K+G KV
Sbjct 80 IVLKAQILAGGRGKGVFN-SGLKGGVHLTKDPNVVGQLAKQMIGYNLATKQTPKEGVKVN 138
Query 91 RLLI 94
++++
Sbjct 139 KVMV 142
> xla:447333 sucla2, MGC82958; succinate-CoA ligase, ADP-forming,
beta subunit (EC:6.2.1.5); K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=458
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/64 (51%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
LVVKAQ+LAGGR G F E G KGGV +P+ +A+ MI K L TKQT +KGR
Sbjct 90 LVVKAQVLAGGRGKGTF-EGGLKGGVKIVYSPEEAKDIASQMIGKKLFTKQTGEKGRICN 148
Query 91 RLLI 94
+ I
Sbjct 149 HVFI 152
> hsa:8803 SUCLA2, A-BETA, MTDPS5, SCS-betaA; succinate-CoA ligase,
ADP-forming, beta subunit (EC:6.2.1.5); K01900 succinyl-CoA
synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=463
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/64 (48%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQ+LAGGR G F E+G KGGV +P+ +V++ MI K L TKQT +KGR
Sbjct 95 VVIKAQVLAGGRGKGTF-ESGLKGGVKIVFSPEEAKAVSSQMIGKKLFTKQTGEKGRICN 153
Query 91 RLLI 94
++L+
Sbjct 154 QVLV 157
> dre:317746 suclg2, cb625, wu:fd44e06; succinate-CoA ligase,
GDP-forming, beta subunit; K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=432
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 30/64 (46%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQILAGGR G F +G KGGVH +P +V +A+ M+ L TKQT K+G +V+
Sbjct 80 IVLKAQILAGGRGKGVFN-SGLKGGVHLTKDPAVVGELASKMLGYNLTTKQTPKEGVEVK 138
Query 91 RLLI 94
+++
Sbjct 139 TVMV 142
> sce:YGR244C LSC2; Beta subunit of succinyl-CoA ligase, which
is a mitochondrial enzyme of the TCA cycle that catalyzes the
nucleotide-dependent conversion of succinyl-CoA to succinate
(EC:6.2.1.4); K01900 succinyl-CoA synthetase beta subunit
[EC:6.2.1.4 6.2.1.5]
Length=427
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 32/66 (48%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 29 NALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRK 88
N LV+KAQ L GGR G F + GYK GVH +P+ VA M+N LITKQT G+
Sbjct 71 NKLVIKAQALTGGRGKGHF-DTGYKSGVHMIESPQQAEDVAKEMLNHNLITKQTGIAGKP 129
Query 89 VRRLLI 94
V + I
Sbjct 130 VSAVYI 135
> mmu:20916 Sucla2, 4930547K18Rik; succinate-Coenzyme A ligase,
ADP-forming, beta subunit (EC:6.2.1.5); K01900 succinyl-CoA
synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=463
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQ+LAGGR G F +G KGGV +P+ +V++ MI + LITKQT +KGR
Sbjct 95 VVIKAQVLAGGRGKGTFT-SGLKGGVKIVFSPEEAKAVSSQMIGQKLITKQTGEKGRICN 153
Query 91 RLLI 94
++L+
Sbjct 154 QVLV 157
> dre:406299 sucla2, wu:fb18c11, wu:fj38c11, wu:fj90d07, zgc:73397;
succinate-CoA ligase, ADP-forming, beta subunit (EC:6.2.1.5);
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=466
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
LV+KAQ+LAGGR G F E G KGGV +P+ +++ MI K L TKQT + GR
Sbjct 98 LVIKAQVLAGGRGKGTF-EGGLKGGVRIVYSPEEARDISSKMIGKKLFTKQTGEAGRICN 156
Query 91 RLLI 94
++ +
Sbjct 157 QVFV 160
> xla:734975 suclg2, MGC132166, gbeta; succinate-CoA ligase, GDP-forming,
beta subunit; K01900 succinyl-CoA synthetase beta
subunit [EC:6.2.1.4 6.2.1.5]
Length=431
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 41/64 (64%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQILAGGR G F ++G KGGVH +P V + MI L TKQT G KV+
Sbjct 79 IVLKAQILAGGRGKGTF-DSGLKGGVHLTKDPSKVEELTKQMIGFNLTTKQTAAGGVKVQ 137
Query 91 RLLI 94
++++
Sbjct 138 KVMV 141
> ath:AT2G20420 succinyl-CoA ligase (GDP-forming) beta-chain,
mitochondrial, putative / succinyl-CoA synthetase, beta chain,
putative / SCS-beta, putative (EC:6.2.1.5 6.2.1.4); K01900
succinyl-CoA synthetase beta subunit [EC:6.2.1.4 6.2.1.5]
Length=421
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 28 ENALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGR 87
E+ LVVK+QILAGGR LG FK +G KGGVH + +A M+ + L+TKQT +G+
Sbjct 68 ESELVVKSQILAGGRGLGTFK-SGLKGGVH-IVKRDEAEEIAGKMLGQVLVTKQTGPQGK 125
Query 88 KVRRLLI 94
V ++ +
Sbjct 126 VVSKVYL 132
> cel:F47B10.1 hypothetical protein; K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=435
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 32 VVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVRR 91
VVKAQ+LAGGR G F +G +GGV P V A MI LITKQT +G+K
Sbjct 67 VVKAQVLAGGRGKGRFS-SGLQGGVQIVFTPDEVKQKAGMMIGANLITKQTDHRGKKCEE 125
Query 92 LLI 94
+++
Sbjct 126 VMV 128
> cel:C50F7.4 hypothetical protein; K01900 succinyl-CoA synthetase
beta subunit [EC:6.2.1.4 6.2.1.5]
Length=415
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 28 ENALVVKAQILAGGRSLGFFKENGYKG--GVHACINPKLVFSVANNMINKTLITKQTTKK 85
++ VVKAQILAGGR G F NG KG GV + MI K L+TKQTT +
Sbjct 59 DHEYVVKAQILAGGRGKGKFI-NGTKGIGGVFITKEKDAALEAIDEMIGKRLVTKQTTSE 117
Query 86 GRKVRRLLI 94
G +V +++I
Sbjct 118 GVRVDKVMI 126
> tpv:TP01_0677 ATP-specific succinyl-CoA synthetase beta subunit;
K01900 succinyl-CoA synthetase beta subunit [EC:6.2.1.4
6.2.1.5]
Length=453
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
LVVKA +L GGR G F G+K GV +P V + A M+ L TKQT KG
Sbjct 76 LVVKALVLTGGRGKGTFN-TGFK-GVEIVKSPSEVSACARGMLGNYLTTKQTVGKGLLCT 133
Query 91 RLLI 94
+L+
Sbjct 134 EVLV 137
> bbo:BBOV_IV010490 23.m06275; succinly CoA-ligase beta subunit
(EC:6.2.1.5); K01900 succinyl-CoA synthetase beta subunit
[EC:6.2.1.4 6.2.1.5]
Length=436
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 34/64 (53%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KA +L GGR G F G V +P+ ++A MI L+TKQT G K +
Sbjct 85 VVLKALVLTGGRGKGKFVGTDISG-VELAKSPERAKTLAEGMIGNVLVTKQTGASGLKCK 143
Query 91 RLLI 94
+L+
Sbjct 144 EVLV 147
> eco:b0728 sucC, ECK0716, JW0717; succinyl-CoA synthetase, beta
subunit (EC:6.2.1.5); K01903 succinyl-CoA synthetase beta
subunit [EC:6.2.1.5]
Length=388
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 8/63 (12%)
Query 32 VVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVRR 91
VVK Q+ AGGR G GGV + + + + A N + K L+T QT G+ V +
Sbjct 44 VVKCQVHAGGR--------GKAGGVKVVNSKEDIRAFAENWLGKRLVTYQTDANGQPVNQ 95
Query 92 LLI 94
+L+
Sbjct 96 ILV 98
Lambda K H
0.316 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069995292
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40