bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1517_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
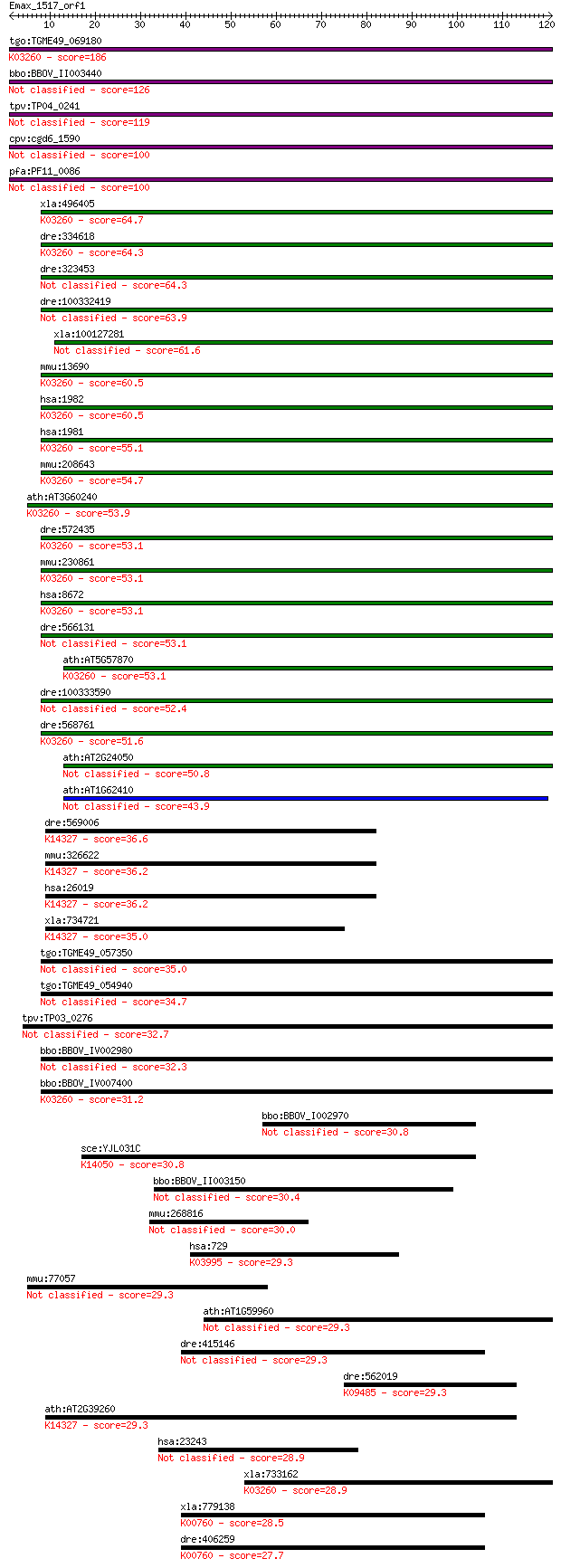
tgo:TGME49_069180 MIF4G domain-containing protein (EC:3.1.1.32... 186 2e-47
bbo:BBOV_II003440 18.m06288; MIF4G domain containing protein 126 2e-29
tpv:TP04_0241 hypothetical protein 119 3e-27
cpv:cgd6_1590 eukaryotic translation initiation factor 4 gamma... 100 1e-21
pfa:PF11_0086 MIF4G domain containing protein 100 1e-21
xla:496405 nat1, DAP5, EIF4G2, p97; eIF4G-related protein NAT1... 64.7 7e-11
dre:334618 eif4g2b, NAT1B, eif4g2, hm:zeh1307, wu:fa14h01, wu:... 64.3 8e-11
dre:323453 eif4g2a, MGC192940, NAT1A, im:7148615, wu:fb81f07, ... 64.3 9e-11
dre:100332419 eukaryotic translation initiation factor 4, gamm... 63.9 1e-10
xla:100127281 eif4g2, dap5, nat1, p97; eukaryotic translation ... 61.6 5e-10
mmu:13690 Eif4g2, AA589388, DAP-5, E130105L11Rik, Nat1, Natm1,... 60.5 1e-09
hsa:1982 EIF4G2, AAG1, DAP5, FLJ41344, NAT1, P97; eukaryotic t... 60.5 1e-09
hsa:1981 EIF4G1, DKFZp686A1451, EIF-4G1, EIF4F, EIF4G, EIF4GI,... 55.1 5e-08
mmu:208643 Eif4g1, E030015G23Rik, MGC37551, MGC90776, eIF4GI; ... 54.7 6e-08
ath:AT3G60240 EIF4G; EIF4G (EUKARYOTIC TRANSLATION INITIATION ... 53.9 1e-07
dre:572435 MGC158450, wu:fb50a05, wu:fc60b06; zgc:158450; K032... 53.1 2e-07
mmu:230861 Eif4g3, 1500002J22Rik, 4833436O05, 4930523M17Rik, G... 53.1 2e-07
hsa:8672 EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII; eukaryotic transl... 53.1 2e-07
dre:566131 eukaryotic translation initiation factor 4 gamma, 3... 53.1 2e-07
ath:AT5G57870 eukaryotic translation initiation factor 4F, put... 53.1 2e-07
dre:100333590 eukaryotic translation initiation factor 4 gamma... 52.4 3e-07
dre:568761 si:dkey-1a7.2; K03260 translation initiation factor 4G 51.6 6e-07
ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-con... 50.8 1e-06
ath:AT1G62410 MIF4G domain-containing protein 43.9 1e-04
dre:569006 upf2; UPF2 regulator of nonsense transcripts homolo... 36.6 0.020
mmu:326622 Upf2; UPF2 regulator of nonsense transcripts homolo... 36.2 0.026
hsa:26019 UPF2, DKFZp434D222, HUPF2, KIAA1408, MGC138834, MGC1... 36.2 0.026
xla:734721 upf2, hupf2, rent2, smg-3; UPF2 regulator of nonsen... 35.0 0.052
tgo:TGME49_057350 eukaryotic translation initiation factor, pu... 35.0 0.058
tgo:TGME49_054940 hypothetical protein 34.7 0.077
tpv:TP03_0276 hypothetical protein 32.7 0.25
bbo:BBOV_IV002980 21.m02788; eukaryotic initiation factor 4G m... 32.3 0.37
bbo:BBOV_IV007400 23.m06334; MIF4G domain containing protein; ... 31.2 0.90
bbo:BBOV_I002970 19.m02223; phosphatidylinositol 3- and 4-kina... 30.8 1.1
sce:YJL031C BET4; Alpha subunit of Type II geranylgeranyltrans... 30.8 1.2
bbo:BBOV_II003150 18.m06263; RNA pseudouridine synthase A 2 30.4
mmu:268816 Gm628; predicted gene 628 30.0
hsa:729 C6; complement component 6; K03995 complement component 6 29.3
mmu:77057 Ston1, 4921524J06Rik, C86231, Salf, Sblf; stonin 1 29.3
ath:AT1G59960 aldo/keto reductase, putative 29.3 3.1
dre:415146 hprt1l, zgc:86643; hypoxanthine phosphoribosyltrans... 29.3 3.1
dre:562019 fc07b10; wu:fc07b10; K09485 heat shock protein 110kDa 29.3 3.4
ath:AT2G39260 RNA binding / binding / protein binding; K14327 ... 29.3 3.4
hsa:23243 ANKRD28, KIAA0379, PITK; ankyrin repeat domain 28 28.9
xla:733162 eif4g1, eif4g; eukaryotic translation initiation fa... 28.9 4.2
xla:779138 hprt1, MGC82603, hgprt, hprt, prtfdc1; hypoxanthine... 28.5 5.9
dre:406259 hprt1, id:ibd1344, id:ibd5108, wu:fc10g09, zgc:5622... 27.7 8.3
> tgo:TGME49_069180 MIF4G domain-containing protein (EC:3.1.1.32
3.4.21.72); K03260 translation initiation factor 4G
Length=2668
Score = 186 bits (472), Expect = 2e-47, Method: Composition-based stats.
Identities = 82/120 (68%), Positives = 94/120 (78%), Gaps = 0/120 (0%)
Query 1 YEFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
+EF YK M GNMIFVGELLKS+MIS ILLECIDRLLQKR ECI S +DQG+ H+
Sbjct 2202 FEFAQLYKARMKGNMIFVGELLKSRMISHRILLECIDRLLQKRLECIDISGGEDQGVPHM 2261
Query 61 EALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
EALCAFLHTVGPFF+NP+W+ Y EFC R+ +V L D SLPFRVRCL+ DVLD+RA W
Sbjct 2262 EALCAFLHTVGPFFENPKWKFYEEFCERIKVVQKLQKDESLPFRVRCLLKDVLDNRAEKW 2321
> bbo:BBOV_II003440 18.m06288; MIF4G domain containing protein
Length=1043
Score = 126 bits (317), Expect = 2e-29, Method: Composition-based stats.
Identities = 53/120 (44%), Positives = 78/120 (65%), Gaps = 0/120 (0%)
Query 1 YEFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
+EFEL YK M GNMIFVGEL K K+++ +L+ C+ + KR ECIAA+ + G +H+
Sbjct 707 FEFELKYKHRMRGNMIFVGELFKQKLLAAKLLITCLSEVFAKRNECIAATGRIETGDNHL 766
Query 61 EALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
E +C L TVG FD +W+ GEF + + ++ L + ++ FR+RCL+ +VLDSR W
Sbjct 767 EGMCTLLQTVGKCFDTDKWKYSGEFEKYIQMLTDLGRNPNICFRIRCLIQNVLDSRHGNW 826
> tpv:TP04_0241 hypothetical protein
Length=1429
Score = 119 bits (298), Expect = 3e-27, Method: Composition-based stats.
Identities = 51/120 (42%), Positives = 76/120 (63%), Gaps = 0/120 (0%)
Query 1 YEFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
+E+E YK M GNMIFVGEL K K+++ +L+ C+D++ KR ECI + + G +H+
Sbjct 826 FEYEQKYKHKMRGNMIFVGELFKQKLLAAKLLITCLDQVFLKREECILLYDDVNMGNNHL 885
Query 61 EALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
EA+C L TVG FD +W+ EF +R+ + L + + FR+RCL+ +VLDSR W
Sbjct 886 EAMCTLLQTVGRSFDTNRWKHLSEFEKRIQHLEDLGKNEEISFRIRCLIKNVLDSRMDHW 945
> cpv:cgd6_1590 eukaryotic translation initiation factor 4 gamma;
Nic domain containing protein
Length=1184
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 45/120 (37%), Positives = 70/120 (58%), Gaps = 12/120 (10%)
Query 1 YEFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
+E++L YK M GNMIF+ L++ K+I+ ++L C++ LLQ HH+
Sbjct 808 FEWQLKYKNKMKGNMIFMASLVRKKVIASTVVLMCMEELLQFHLP------------HHL 855
Query 61 EALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
EALC FLH VGPF D+ +W+ Y +F +++ +P R+R L++DV+DSR W
Sbjct 856 EALCVFLHHVGPFLDSERWKHYEDFNTLFLQFEEFSTNKEVPIRIRFLINDVIDSRKNNW 915
> pfa:PF11_0086 MIF4G domain containing protein
Length=3334
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 50/120 (41%), Positives = 73/120 (60%), Gaps = 0/120 (0%)
Query 1 YEFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
+EFE +K + GNM+FVGEL+KS +IS I+ CI +LL+KR I+ N +G H+
Sbjct 2798 FEFEQMHKNKVRGNMLFVGELVKSGIISIPIVFVCIKQLLEKRESYISIKNDTKEGNLHL 2857
Query 61 EALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
EALC FL+TVG D + + + + L ++ S+ FRVRCL+ DV+D+R W
Sbjct 2858 EALCMFLNTVGEILDTHEKANQEKVKELYNTLNELVNNESITFRVRCLIKDVIDNRNEKW 2917
> xla:496405 nat1, DAP5, EIF4G2, p97; eIF4G-related protein NAT1;
K03260 translation initiation factor 4G
Length=903
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL CI LL+K+ KD G +E LC +
Sbjct 209 KIKMLGNIKFIGELGKLDLIHESILHRCIKALLEKKKRV----QLKDMG-EDLECLCQIM 263
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM +L ++ LP R+R L+ D ++ RA W
Sbjct 264 RTVGPRLDHEKAKSLMDQYFARM---CALKANKELPARIRFLLQDTMELRANNW 314
> dre:334618 eif4g2b, NAT1B, eif4g2, hm:zeh1307, wu:fa14h01, wu:fb44h01,
wu:fb59c06, wu:fc21b05; eukaryotic translation initiation
factor 4, gamma 2b; K03260 translation initiation factor
4G
Length=898
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 199 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 253
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL ++ LP R+R L+ D ++ R W
Sbjct 254 RTVGPRLDHEKAKSLMDQYFGRM---QSLMNNKDLPARIRFLLQDTVELRENNW 304
> dre:323453 eif4g2a, MGC192940, NAT1A, im:7148615, wu:fb81f07,
wu:fb82b06, wu:fb98h08; eukaryotic translation initiation
factor 4, gamma 2a
Length=891
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 199 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 253
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL ++ LP R+R L+ D ++ R W
Sbjct 254 RTVGPRLDHEKAKSLMDQYFGRM---RSLMNNKDLPARIRFLLQDTVELRENNW 304
> dre:100332419 eukaryotic translation initiation factor 4, gamma
2a-like
Length=953
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 271 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 325
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL ++ LP R+R L+ D ++ R W
Sbjct 326 RTVGPRLDHEKAKSLMDQYFGRM---RSLMNNKDLPARIRFLLQDTVELRENNW 376
> xla:100127281 eif4g2, dap5, nat1, p97; eukaryotic translation
initiation factor 4 gamma, 2
Length=691
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 59/111 (53%), Gaps = 9/111 (8%)
Query 11 MTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTV 70
M GN+ F+GEL K +I ++IL CI LL+K+ KD G +E LC + TV
Sbjct 1 MLGNIKFIGELGKLDLIHESILHRCIKALLEKKKRV----QLKDMG-EDLECLCQIMRTV 55
Query 71 GPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
GP D+ + + L ++ RM +L + LP R+R L+ D ++ R W
Sbjct 56 GPRLDHEKAKSLMDQYFARM---CALKTSKELPARIRFLLQDTMELRGNHW 103
> mmu:13690 Eif4g2, AA589388, DAP-5, E130105L11Rik, Nat1, Natm1,
p97; eukaryotic translation initiation factor 4, gamma 2;
K03260 translation initiation factor 4G
Length=868
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 60/114 (52%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 204 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 258
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL LP R+R L+ D ++ R W
Sbjct 259 RTVGPRLDHERAKSLMDQYFARM---CSLMLSKELPARIRFLLQDTVELREHHW 309
> hsa:1982 EIF4G2, AAG1, DAP5, FLJ41344, NAT1, P97; eukaryotic
translation initiation factor 4 gamma, 2; K03260 translation
initiation factor 4G
Length=869
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 60/114 (52%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 204 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 258
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL LP R+R L+ D ++ R W
Sbjct 259 RTVGPRLDHERAKSLMDQYFARM---CSLMLSKELPARIRFLLQDTVELREHHW 309
> hsa:1981 EIF4G1, DKFZp686A1451, EIF-4G1, EIF4F, EIF4G, EIF4GI,
P220; eukaryotic translation initiation factor 4 gamma, 1;
K03260 translation initiation factor 4G
Length=1606
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 58/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 899 RRRSLGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 946
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DVLD R + W
Sbjct 947 TTIGKDLDFEKAKPRMDQYFNQMEKIIKEKKTSS---RIRFMLQDVLDLRGSNW 997
> mmu:208643 Eif4g1, E030015G23Rik, MGC37551, MGC90776, eIF4GI;
eukaryotic translation initiation factor 4, gamma 1; K03260
translation initiation factor 4G
Length=1593
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 58/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 889 RRRSLGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 936
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DVLD R + W
Sbjct 937 TTIGKDLDFAKAKPRMDQYFNQMEKIIKEKKTSS---RIRFMLQDVLDLRQSNW 987
> ath:AT3G60240 EIF4G; EIF4G (EUKARYOTIC TRANSLATION INITIATION
FACTOR 4G); translation initiation factor; K03260 translation
initiation factor 4G
Length=1723
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 61/118 (51%), Gaps = 15/118 (12%)
Query 5 LNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALC 64
L ++ M GN+ +GEL K +M+++ I+ CI +LL ++D ++EALC
Sbjct 1212 LQVRRRMLGNIRLIGELYKKRMLTEKIMHACIQKLL---------GYNQDPHEENIEALC 1262
Query 65 AFLHTVGPFFDN--PQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
+ T+G D+ ++++ G F +M + L+ L RVR ++ + +D R W
Sbjct 1263 KLMSTIGVMIDHNKAKFQMDGYF-EKMKM---LSCKQELSSRVRFMLINAIDLRKNKW 1316
> dre:572435 MGC158450, wu:fb50a05, wu:fc60b06; zgc:158450; K03260
translation initiation factor 4G
Length=1585
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 34/114 (29%), Positives = 56/114 (49%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
+K GN+ F+GEL K KM+++ I+ +CI +LL+ E +E LC L
Sbjct 860 RKRSLGNIKFIGELFKLKMLTEPIMHDCIVKLLKNHDE------------DSLECLCRLL 907
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DVLD R W
Sbjct 908 STIGKDLDFEKAKPRMDQYFHQMEKIIKEKKTSS---RIRFMLQDVLDLRKNNW 958
> mmu:230861 Eif4g3, 1500002J22Rik, 4833436O05, 4930523M17Rik,
G1-419-52, eIF4GII, repro8; eukaryotic translation initiation
factor 4 gamma, 3; K03260 translation initiation factor 4G
Length=1578
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 57/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 899 RRRSIGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 946
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DV+D R W
Sbjct 947 TTIGKDLDFEKAKPRMDQYFNQMEKIVKERKTSS---RIRFMLQDVIDLRLCNW 997
> hsa:8672 EIF4G3, eIF-4G_3, eIF4G_3, eIF4GII; eukaryotic translation
initiation factor 4 gamma, 3; K03260 translation initiation
factor 4G
Length=1621
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 57/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 922 RRRSIGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 969
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DV+D R W
Sbjct 970 TTIGKDLDFEKAKPRMDQYFNQMEKIVKERKTSS---RIRFMLQDVIDLRLCNW 1020
> dre:566131 eukaryotic translation initiation factor 4 gamma,
3-like
Length=1697
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 59/114 (51%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ TGN+ F+GEL K KM+++ I+ +C+ +LL+ N D+ + E LC L
Sbjct 996 RRRSTGNIKFIGELFKLKMLTEPIMHDCVVKLLK---------NHDDESL---ECLCRLL 1043
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DV+D R W
Sbjct 1044 TTIGKDLDFEKAKPRMDQYFNQMEKIVKERKTSS---RIRFMLQDVIDLRLHNW 1094
> ath:AT5G57870 eukaryotic translation initiation factor 4F, putative
/ eIF-4F, putative; K03260 translation initiation factor
4G
Length=776
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 56/111 (50%), Gaps = 14/111 (12%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAE-CIAASNSKDQGIHHVEALCAFLHTVG 71
GN+ +GELLK KM+ + I+ + LL + C A N VEA+C F T+G
Sbjct 339 GNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEEN--------VEAICHFFKTIG 390
Query 72 PFFDN--PQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
D R+ + +R+ +L+ + L R+R ++ +++D R+ GW
Sbjct 391 KQLDGNVKSKRINDVYFKRLQ---ALSKNPQLELRLRFMVQNIIDMRSNGW 438
> dre:100333590 eukaryotic translation initiation factor 4 gamma,
1-like
Length=966
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 57/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++ I+ +CI +LL+ E +E LC L
Sbjct 694 RRRSLGNIKFIGELFKLKMLTENIMHDCIVKLLKNHDE------------ESLECLCRLL 741
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DVLD R + W
Sbjct 742 ATIGKDLDFEKAKPRMDQYFNQMEKIIKERKTSS---RIRFMLQDVLDLRRSNW 792
> dre:568761 si:dkey-1a7.2; K03260 translation initiation factor
4G
Length=1535
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 57/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K +M+++AI+ +C+ +LL+ E +E LC L
Sbjct 836 RRRSIGNIKFIGELFKLRMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 883
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DV+D R W
Sbjct 884 TTIGKDLDFEKAKPRMDQYFNQMEKIVKERKTSS---RIRFMLQDVIDLRLHNW 934
> ath:AT2G24050 MIF4G domain-containing protein / MA3 domain-containing
protein
Length=747
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 53/111 (47%), Gaps = 14/111 (12%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGP 72
GN+ +GELLK KM+ + I+ + LL + A VEALC F T+G
Sbjct 302 GNIRLIGELLKQKMVPEKIVHHIVQELLGDDTKACPAEGD-------VEALCQFFITIGK 354
Query 73 FFDN-PQWRLYGE--FCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
D+ P+ R + F R + LA L R+R ++ +V+D RA W
Sbjct 355 QLDDSPRSRGINDTYFGR----LKELARHPQLELRLRFMVQNVVDLRANKW 401
> ath:AT1G62410 MIF4G domain-containing protein
Length=223
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 55/107 (51%), Gaps = 10/107 (9%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGP 72
GN+ F GEL +M+++ ++L +LL+ AE + S K + A+C FL+TVG
Sbjct 104 GNLRFCGELFLKRMLTEKVVLAIGQKLLED-AEQMCPSEEK------IIAICLFLNTVGK 156
Query 73 FFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAG 119
D+ +L E RR+ +L++ L +R ++ ++ + G
Sbjct 157 KLDSLNSKLMNEILRRL---KNLSNHPQLVMSLRLMVGKIIHLHSIG 200
> dre:569006 upf2; UPF2 regulator of nonsense transcripts homolog
(yeast); K14327 regulator of nonsense transcripts 2
Length=594
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 32/74 (43%), Gaps = 13/74 (17%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLH 68
+T + F+GEL K KM S+ L C+ LL D HH+E C L
Sbjct 245 ETKNKTVRFIGELAKFKMFSKTDTLHCLKMLLS------------DFSHHHIEMACTLLE 292
Query 69 TVGPF-FDNPQWRL 81
T G F F +P L
Sbjct 293 TCGRFLFRSPDSHL 306
> mmu:326622 Upf2; UPF2 regulator of nonsense transcripts homolog
(yeast); K14327 regulator of nonsense transcripts 2
Length=1269
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 33/74 (44%), Gaps = 13/74 (17%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLH 68
+T + F+GEL K KM ++ L C+ LL D HH+E C L
Sbjct 659 ETKNKTVRFIGELTKFKMFTKNDTLHCLKMLLS------------DFSHHHIEMACTLLE 706
Query 69 TVGPF-FDNPQWRL 81
T G F F +P+ L
Sbjct 707 TCGRFLFRSPESHL 720
> hsa:26019 UPF2, DKFZp434D222, HUPF2, KIAA1408, MGC138834, MGC138835,
RENT2, smg-3; UPF2 regulator of nonsense transcripts
homolog (yeast); K14327 regulator of nonsense transcripts
2
Length=1272
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 33/74 (44%), Gaps = 13/74 (17%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLH 68
+T + F+GEL K KM ++ L C+ LL D HH+E C L
Sbjct 661 ETKNKTVRFIGELTKFKMFTKNDTLHCLKMLLS------------DFSHHHIEMACTLLE 708
Query 69 TVGPF-FDNPQWRL 81
T G F F +P+ L
Sbjct 709 TCGRFLFRSPESHL 722
> xla:734721 upf2, hupf2, rent2, smg-3; UPF2 regulator of nonsense
transcripts homolog; K14327 regulator of nonsense transcripts
2
Length=1264
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 28/66 (42%), Gaps = 12/66 (18%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLH 68
+T + F+GEL K KM ++ L C+ LL D HH+E C L
Sbjct 653 ETKNKTVRFIGELAKFKMFNKTDTLHCLKMLLS------------DFSHHHIEMACTLLE 700
Query 69 TVGPFF 74
T G F
Sbjct 701 TCGRFL 706
> tgo:TGME49_057350 eukaryotic translation initiation factor,
putative
Length=820
Score = 35.0 bits (79), Expect = 0.058, Method: Composition-based stats.
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 16/116 (13%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K + G + +GEL + K++ I+ + + L+ K E D+ H +E +
Sbjct 628 KNRILGVVKLIGELFQRKILGFPIVRDVVVDLVIKNEE-------PDE--HFIECFVQLI 678
Query 68 HTVGPFFD-NPQWR--LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T G + D NP+ + L F R + L R++C++ D LD R A W
Sbjct 679 ATTGYYIDQNPKVKAVLDSWFGR----LTELQKKPCYSKRLKCIIQDTLDMRKAEW 730
> tgo:TGME49_054940 hypothetical protein
Length=3799
Score = 34.7 bits (78), Expect = 0.077, Method: Composition-based stats.
Identities = 34/119 (28%), Positives = 52/119 (43%), Gaps = 17/119 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGI----HHVEAL 63
K + GNM F+GEL K I+ ++L + L+ +S D + H VE L
Sbjct 3229 KDRVLGNMRFIGELYLRKCIAPSVLKAVVTSLV--------FGDSGDPDVYPDEHFVECL 3280
Query 64 CAFLHTVGPFFD-NPQ-WRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
L T+G + PQ ++ EF M + L ++ R+ + DVLD R W
Sbjct 3281 TELLITIGFTLEQQPQSQQMLHEF---MGKLQDLQQKANYSKRIIYKIQDVLDLRTRNW 3336
> tpv:TP03_0276 hypothetical protein
Length=359
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 54/123 (43%), Gaps = 20/123 (16%)
Query 4 ELNYKKTMT---GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHV 60
E++ KK T G + +GEL + KM+ I+ + + L+ +++ H +
Sbjct 178 EVSVKKLKTRTLGTVKMIGELFQRKMLGFKIVNKVVFDLVM----------NQEPHEHLI 227
Query 61 EALCAFLHTVGPFFD-NPQWRLYGE--FCRRMDLVASLASDSSLPFRVRCLMSDVLDSRA 117
E ++ G + D NP R + F R + LA R++CL+ DVL+
Sbjct 228 ECFIQLIYGTGYYIDKNPNLRPVLDLWFGR----LKELAQRKEYSKRIKCLIQDVLNLPK 283
Query 118 AGW 120
A W
Sbjct 284 AQW 286
> bbo:BBOV_IV002980 21.m02788; eukaryotic initiation factor 4G
middle domain containing protein
Length=1481
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 30/115 (26%), Positives = 53/115 (46%), Gaps = 10/115 (8%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
KK + GN+ F+GEL K+IS IL + LLQ + + S + VE+ +
Sbjct 1067 KKWVLGNIRFMGELFLRKVISVGILKRIVCTLLQ-----MDSDGSHIPCEYLVESFLELI 1121
Query 68 HTVGPFFDN-PQW-RLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G + P + ++ M + +L + + R+ + D++D RA W
Sbjct 1122 TTIGYTLEQMPHGPDMLNDY---MGHLTNLKKNGNYSLRIVYKIQDLIDLRAKNW 1173
> bbo:BBOV_IV007400 23.m06334; MIF4G domain containing protein;
K03260 translation initiation factor 4G
Length=367
Score = 31.2 bits (69), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 52/116 (44%), Gaps = 16/116 (13%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K + G + +GEL + +++ I+ + + L+ I+A + H +E +
Sbjct 192 KTRILGTVKMIGELFQRRILGFKIVNQVVLDLV------ISADEPHE---HLIECFLQLI 242
Query 68 HTVGPFFD-NPQWR--LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
++ G + D NP R L F R +L+ R++C+M DVLD A W
Sbjct 243 YSTGYYIDRNPSLRPVLDMWFGRLKELMLKRCYSK----RIKCVMQDVLDLPKAQW 294
> bbo:BBOV_I002970 19.m02223; phosphatidylinositol 3- and 4-kinase
domain containing protein (EC:2.7.1.-)
Length=1151
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 57 IHHVEALCAFLHTVGP-----FFDNPQWRLYGEFCRRMDLVASLASDSSLPF 103
+ +AL L+ G F DNP + EFCRR++ V SLA+ S++ +
Sbjct 954 VDETKALSDILNDYGSIGGYVFEDNPS--AFEEFCRRLNFVGSLAAYSAITY 1003
> sce:YJL031C BET4; Alpha subunit of Type II geranylgeranyltransferase
required for vesicular transport between the endoplasmic
reticulum and the Golgi; provides a membrane attachment
moiety to Rab-like proteins Ypt1p and Sec4p (EC:2.5.1.60);
K14050 geranylgeranyl transferase type-2 subunit alpha [EC:2.5.1.60]
Length=327
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 40/87 (45%), Gaps = 11/87 (12%)
Query 17 FVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDN 76
+ ELL+ K + ++ R+ N +D+ I+ +EAL T N
Sbjct 9 WTKELLRQKRVQDE------KKIYDYRSLTENVLNMRDEKIYSIEALKK---TSELLEKN 59
Query 77 PQWRLYGEFCRRMDLVASLASDSSLPF 103
P++ + R D++ASLAS+ +PF
Sbjct 60 PEFNAIWNY--RRDIIASLASELEIPF 84
> bbo:BBOV_II003150 18.m06263; RNA pseudouridine synthase A 2
Length=295
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 6/66 (9%)
Query 33 LECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLV 92
+ C D L +R + AS+ D+G+H E C +L F P + G+ MD V
Sbjct 143 VHCPDTLPPERRFTLIASSRTDKGVHATETACQYL----SFDKEPPYN--GDIDAIMDKV 196
Query 93 ASLASD 98
L D
Sbjct 197 NRLLPD 202
> mmu:268816 Gm628; predicted gene 628
Length=424
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 32 LLECIDRLLQKRAECIAASNSKDQGIHHVEALCAF 66
LLEC+ L++K + ++ + Q IH V +LCA
Sbjct 284 LLECLMTLMEKEPQDTLVTSIRQQAIHIVSSLCAL 318
> hsa:729 C6; complement component 6; K03995 complement component
6
Length=934
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 1/46 (2%)
Query 41 QKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFC 86
Q +ECI S +D HH E LC F +F +P + E C
Sbjct 779 QSGSECICMSPEEDCS-HHSEDLCVFDTDSNDYFTSPACKFLAEKC 823
> mmu:77057 Ston1, 4921524J06Rik, C86231, Salf, Sblf; stonin 1
Length=730
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 32/64 (50%), Gaps = 11/64 (17%)
Query 5 LNYKKTMTGNMIFVGELLKSKMISQAILL-------ECI----DRLLQKRAECIAASNSK 53
L+ + ++ G + G+L++S +++Q L EC DR LQKR EC +
Sbjct 412 LDIQDSLWGKVTKEGQLVESAVVTQICCLCFLNGPAECFLALNDRELQKRDECYFEKEPE 471
Query 54 DQGI 57
+GI
Sbjct 472 KKGI 475
> ath:AT1G59960 aldo/keto reductase, putative
Length=326
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 32/79 (40%), Gaps = 16/79 (20%)
Query 44 AECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQW--RLYGEFCRRMDLVASLASDSSL 101
A+CI SN + + H+ ++ +V +P W R E CR D+V + S
Sbjct 163 AKCIGVSNFSCKKLQHILSIATIPPSVNQVEMSPIWQQRKLRELCRSNDIVVTAYS---- 218
Query 102 PFRVRCLMSDVLDSRAAGW 120
VL SR A W
Sbjct 219 ----------VLGSRGAFW 227
> dre:415146 hprt1l, zgc:86643; hypoxanthine phosphoribosyltransferase
1, like
Length=215
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 29/69 (42%), Gaps = 14/69 (20%)
Query 39 LLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLA-- 96
L++ R E +A +D G HH+ ALC Y F MD + +L
Sbjct 38 LIKDRTERLARDIVRDMGGHHIVALCVLKGG------------YKFFADLMDFIKTLNQH 85
Query 97 SDSSLPFRV 105
SD S+P V
Sbjct 86 SDKSVPLTV 94
> dre:562019 fc07b10; wu:fc07b10; K09485 heat shock protein 110kDa
Length=826
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 4/40 (10%)
Query 75 DNPQ--WRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDV 112
DNP+ RLY E C ++ + S A+ S LP + C M+D+
Sbjct 258 DNPRALLRLYQE-CEKLKKLMS-ANSSDLPLNIECFMNDI 295
> ath:AT2G39260 RNA binding / binding / protein binding; K14327
regulator of nonsense transcripts 2
Length=1181
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 46/106 (43%), Gaps = 17/106 (16%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHH-VEALCAFL 67
+T N+ F+GEL K K++ ++ C+ L D+ HH ++ C L
Sbjct 559 ETKIRNIRFIGELCKFKIVPAGLVFSCLKACL-------------DEFTHHNIDVACNLL 605
Query 68 HTVGPF-FDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDV 112
T G F + +P+ L +D++ L + +L R L+ +
Sbjct 606 ETCGRFLYRSPETTL--RMTNMLDILMRLKNVKNLDPRQSTLVENA 649
> hsa:23243 ANKRD28, KIAA0379, PITK; ankyrin repeat domain 28
Length=899
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query 34 ECIDRLLQKRAECIAASNSKDQGIH------HVEALCAFLHTVGPFFDNP 77
EC+D LLQ A+C+ + IH H+ L A L + NP
Sbjct 546 ECVDALLQHGAKCLLRDSRGRTPIHLSAACGHIGVLGALLQSAASMDANP 595
> xla:733162 eif4g1, eif4g; eukaryotic translation initiation
factor 4 gamma, 1; K03260 translation initiation factor 4G
Length=686
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query 53 KDQGIHHVEALCAFLHTVGPF--FDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMS 110
K+ +E LC L T+G F+ + R+ ++ +MD + SS R+R ++
Sbjct 10 KNHDEESLECLCRLLSTIGKDLDFERAKPRM-DQYFNQMDKIIKERKTSS---RIRFMLQ 65
Query 111 DVLDSRAAGW 120
DV+D R W
Sbjct 66 DVIDLRLCNW 75
> xla:779138 hprt1, MGC82603, hgprt, hprt, prtfdc1; hypoxanthine
phosphoribosyltransferase 1 (EC:2.4.2.8); K00760 hypoxanthine
phosphoribosyltransferase [EC:2.4.2.8]
Length=216
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 28/69 (40%), Gaps = 14/69 (20%)
Query 39 LLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASL--A 96
L+ R E +A KD G HH+ ALC Y F +D + +L
Sbjct 39 LIMDRTERLARDIMKDMGGHHIVALCVLKGG------------YKFFADLLDYIKALNRN 86
Query 97 SDSSLPFRV 105
SD S+P V
Sbjct 87 SDKSIPMTV 95
> dre:406259 hprt1, id:ibd1344, id:ibd5108, wu:fc10g09, zgc:56221,
zgc:86608; hypoxanthine phosphoribosyltransferase 1 (EC:2.4.2.8);
K00760 hypoxanthine phosphoribosyltransferase [EC:2.4.2.8]
Length=218
Score = 27.7 bits (60), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 28/69 (40%), Gaps = 14/69 (20%)
Query 39 LLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASL--A 96
L+ R E +A KD G HH+ ALC Y F +D + +L
Sbjct 41 LIMDRTERLARDIMKDMGGHHIVALCVLKGG------------YKFFADLLDYIKALNRN 88
Query 97 SDSSLPFRV 105
SD S+P V
Sbjct 89 SDRSIPMTV 97
Lambda K H
0.326 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40