bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
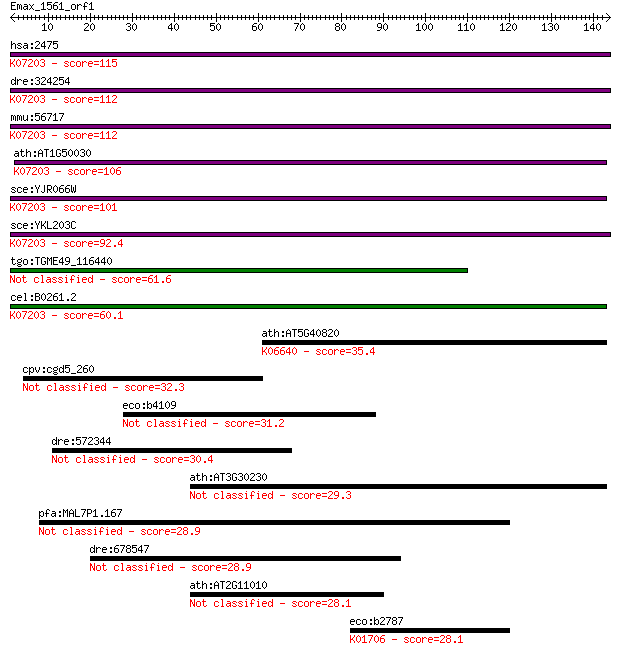
Query= Emax_1561_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
hsa:2475 MTOR, FLJ44809, FRAP, FRAP1, FRAP2, RAFT1, RAPT1; mec... 115 7e-26
dre:324254 mtor, fc22h08, frap1, tor, wu:fc22h08; mechanistic ... 112 3e-25
mmu:56717 Mtor, 2610315D21Rik, AI327068, FRAP, FRAP2, Frap1, M... 112 3e-25
ath:AT1G50030 TOR; TOR (TARGET OF RAPAMYCIN); 1-phosphatidylin... 106 2e-23
sce:YJR066W TOR1, DRR1; PIK-related protein kinase and rapamyc... 101 9e-22
sce:YKL203C TOR2, DRR2; PIK-related protein kinase and rapamyc... 92.4 5e-19
tgo:TGME49_116440 phosphatidylinositol 3- and 4-kinase domain-... 61.6 7e-10
cel:B0261.2 let-363; LEThal family member (let-363); K07203 FK... 60.1 2e-09
ath:AT5G40820 ATRAD3; ATRAD3; binding / inositol or phosphatid... 35.4 0.061
cpv:cgd5_260 hypothetical protein 32.3 0.45
eco:b4109 yjdA, ECK4102, JW4070; mutational suppressor of YhjH... 31.2 1.2
dre:572344 nuclear mitotic apparatus protein 1-like 30.4 1.8
ath:AT3G30230 myosin heavy chain-related 29.3 3.9
pfa:MAL7P1.167 conserved Plasmodium protein, unknown function 28.9 5.4
dre:678547 MGC136845, baat1; zgc:136845 28.9 6.0
ath:AT2G11010 hypothetical protein 28.1 8.8
eco:b2787 gudD, ECK2781, JW2758, ygcX; (D)-glucarate dehydrata... 28.1 9.8
> hsa:2475 MTOR, FLJ44809, FRAP, FRAP1, FRAP2, RAFT1, RAPT1; mechanistic
target of rapamycin (serine/threonine kinase) (EC:2.7.11.1);
K07203 FKBP12-rapamycin complex-associated protein
Length=2549
Score = 115 bits (287), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 64/143 (44%), Positives = 98/143 (68%), Gaps = 1/143 (0%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
AYAKALHYKE EF +G T +L +LI+IN KL Q +AA GVL+Y++K+ ++E++ W+
Sbjct 1392 AYAKALHYKELEFQKGPTPAILESLISINNKLQQPEAAAGVLEYAMKH-FGELEIQATWY 1450
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
EK+ WE A ++Y K + DD E +LG+ RCL+ LGE+ +L E + +++ Q+
Sbjct 1451 EKLHEWEDALVAYDKKMDTNKDDPELMLGRMRCLEALGEWGQLHQQCCEKWTLVNDETQA 1510
Query 121 RMARLAVQASWSLNKWDKMEEFS 143
+MAR+A A+W L +WD MEE++
Sbjct 1511 KMARMAAAAAWGLGQWDSMEEYT 1533
> dre:324254 mtor, fc22h08, frap1, tor, wu:fc22h08; mechanistic
target of rapamycin (serine/threonine kinase); K07203 FKBP12-rapamycin
complex-associated protein
Length=2515
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 65/143 (45%), Positives = 96/143 (67%), Gaps = 1/143 (0%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
AYAKALHYKE EF +G + +L +LI+IN KL Q +AA GVL+Y++K+ ++E++ W+
Sbjct 1375 AYAKALHYKELEFQKGASPLILESLISINNKLQQPEAASGVLEYAMKH-FGELEIQATWY 1433
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
EK+ WE A ++Y K + DD E ILG+ RCL+ LGE+ +L E + E+ Q+
Sbjct 1434 EKLHEWEDALVAYDKKIDMNKDDPELILGRMRCLEALGEWGQLHQQCCEEWTLVSEETQA 1493
Query 121 RMARLAVQASWSLNKWDKMEEFS 143
+MAR+A A+W L WD MEE++
Sbjct 1494 KMARMAAAAAWGLGHWDSMEEYT 1516
> mmu:56717 Mtor, 2610315D21Rik, AI327068, FRAP, FRAP2, Frap1,
MGC118056, RAFT1, RAPT1, flat; mechanistic target of rapamycin
(serine/threonine kinase) (EC:2.7.11.1); K07203 FKBP12-rapamycin
complex-associated protein
Length=2549
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 63/143 (44%), Positives = 98/143 (68%), Gaps = 1/143 (0%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
AYAKALHYKE EF +G T +L +LI+IN KL Q +AA GVL+Y++K+ ++E++ W+
Sbjct 1392 AYAKALHYKELEFQKGPTPAILESLISINNKLQQPEAASGVLEYAMKH-FGELEIQATWY 1450
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
EK+ WE A ++Y K + +D E +LG+ RCL+ LGE+ +L E + +++ Q+
Sbjct 1451 EKLHEWEDALVAYDKKMDTNKEDPELMLGRMRCLEALGEWGQLHQQCCEKWTLVNDETQA 1510
Query 121 RMARLAVQASWSLNKWDKMEEFS 143
+MAR+A A+W L +WD MEE++
Sbjct 1511 KMARMAAAAAWGLGQWDSMEEYT 1533
> ath:AT1G50030 TOR; TOR (TARGET OF RAPAMYCIN); 1-phosphatidylinositol-3-kinase/
protein binding; K07203 FKBP12-rapamycin
complex-associated protein
Length=2454
Score = 106 bits (264), Expect = 2e-23, Method: Composition-based stats.
Identities = 56/151 (37%), Positives = 89/151 (58%), Gaps = 12/151 (7%)
Query 2 YAKALHYKEKEF-------LEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVE 54
+AKALHYKE EF ++ V+ ALI IN +L Q +AA G+L Y+ ++ DV+
Sbjct 1320 FAKALHYKEMEFEGPRSKRMDANPVAVVEALIHINNQLHQHEAAVGILTYAQQHL--DVQ 1377
Query 55 VREKWFEKMGNWELAYLSYKFKHEYKSDD---IEAILGQSRCLDHLGEYERLSTLVDETF 111
++E W+EK+ W+ A +Y K ++ +EA LGQ RCL L +E L+ L E +
Sbjct 1378 LKESWYEKLQRWDDALKAYTLKASQTTNPHLVLEATLGQMRCLAALARWEELNNLCKEYW 1437
Query 112 AQQDEQGQSRMARLAVQASWSLNKWDKMEEF 142
+ + + MA +A QA+W++ +WD+M E+
Sbjct 1438 SPAEPSARLEMAPMAAQAAWNMGEWDQMAEY 1468
> sce:YJR066W TOR1, DRR1; PIK-related protein kinase and rapamycin
target; subunit of TORC1, a complex that controls growth
in response to nutrients by regulating translation, transcription,
ribosome biogenesis, nutrient transport and autophagy;
involved in meiosis (EC:2.7.1.137); K07203 FKBP12-rapamycin
complex-associated protein
Length=2470
Score = 101 bits (251), Expect = 9e-22, Method: Composition-based stats.
Identities = 51/142 (35%), Positives = 84/142 (59%), Gaps = 2/142 (1%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
AYAKALHYKE +F++ + +LI+IN +L Q AA G+L+++ ++ ++++E WF
Sbjct 1341 AYAKALHYKEIKFIKEPENSTIESLISINNQLNQTDAAIGILKHAQQHH--SLQLKETWF 1398
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
EK+ WE A +Y + + + LG+ R L LGE+E+LS L + Q +
Sbjct 1399 EKLERWEDALHAYNEREKAGDTSVSVTLGKMRSLHALGEWEQLSQLAARKWKVSKLQTKK 1458
Query 121 RMARLAVQASWSLNKWDKMEEF 142
+A LA A+W L +WD +E++
Sbjct 1459 LIAPLAAGAAWGLGEWDMLEQY 1480
> sce:YKL203C TOR2, DRR2; PIK-related protein kinase and rapamycin
target; subunit of TORC1, a complex that regulates growth
in response to nutrients and TORC2, a complex that regulates
cell-cycle dependent polarization of the actin cytoskeleton;
involved in meiosis (EC:2.7.11.1 2.7.1.67); K07203 FKBP12-rapamycin
complex-associated protein
Length=2474
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 54/143 (37%), Positives = 89/143 (62%), Gaps = 2/143 (1%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
A+AKALHYKE EFLE + ALI+IN +L Q +A G+L+++ ++ ++++++E W+
Sbjct 1348 AFAKALHYKEVEFLEEPKNSTIEALISINNQLHQTDSAIGILKHAQQH--NELQLKETWY 1405
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
EK+ WE A +Y K D +E ++G+ R L LGE+E LS L E + + +
Sbjct 1406 EKLQRWEDALAAYNEKEAAGEDSVEVMMGKLRSLYALGEWEELSKLASEKWGTAKPEVKK 1465
Query 121 RMARLAVQASWSLNKWDKMEEFS 143
MA LA A+W L +WD++ +++
Sbjct 1466 AMAPLAAGAAWGLEQWDEIAQYT 1488
> tgo:TGME49_116440 phosphatidylinositol 3- and 4-kinase domain-containing
protein (EC:4.1.1.70 2.7.11.1)
Length=2896
Score = 61.6 bits (148), Expect = 7e-10, Method: Composition-based stats.
Identities = 38/109 (34%), Positives = 63/109 (57%), Gaps = 3/109 (2%)
Query 1 AYAKALHYKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
A AKA+ Y+E+ +L V ALI ++ + Q +AA G+L ++ K + V+E W+
Sbjct 966 ANAKAIRYREELWLADPGGSV-EALIRLSHETQQLEAARGILAHAQKKL--RLPVKECWY 1022
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDE 109
++G WE A +Y+ + + E + G+ RCL LGE+ERL+ L D+
Sbjct 1023 VQLGEWEQALEAYEQREREDPSNAEWLKGKMRCLRALGEWERLAFLADD 1071
> cel:B0261.2 let-363; LEThal family member (let-363); K07203
FKBP12-rapamycin complex-associated protein
Length=2695
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 47/188 (25%), Positives = 92/188 (48%), Gaps = 47/188 (25%)
Query 1 AYAKALHYKEKEFLEG----QTT---------DVLGALITINKKLGQEQAAYGVLQYSLK 47
A+AKA YKE L+ QTT + +LIT KL ++ A GV++Y+ +
Sbjct 1450 AFAKACRYKEMSVLKKSGSMQTTFTRKVKLEPNDCQSLITYANKLNVQEEAAGVVRYAER 1509
Query 48 NRISDVEVREKWFEKMGNWELAYLSYKFKHEYKS--------DDIEAILG---------- 89
N + + ++R +W+EK+ WE A +Y+ + + KS D+ + ++
Sbjct 1510 NEM-NFQMRGRWYEKLNEWEKALGAYELEEKKKSSCPNLQVYDEKDHLMTPEEAATAEEA 1568
Query 90 ---QSRCLDHLGEYERLST----LVDETFAQQD--------EQGQSRMARLAVQASWSLN 134
+ RCL+ LG ++ L++ D+ + D +Q +MA +A + +W+++
Sbjct 1569 RMHEMRCLEALGRWDELNSKSVVWADQRGNRNDSVRDEINKKQLDHKMAVIAARGAWAVD 1628
Query 135 KWDKMEEF 142
W++M ++
Sbjct 1629 NWERMADY 1636
> ath:AT5G40820 ATRAD3; ATRAD3; binding / inositol or phosphatidylinositol
kinase/ phosphotransferase, alcohol group as acceptor
/ protein serine/threonine kinase; K06640 ataxia telangiectasia
and Rad3 related [EC:2.7.11.1]
Length=2702
Score = 35.4 bits (80), Expect = 0.061, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 41/82 (50%), Gaps = 1/82 (1%)
Query 61 EKMGNWELAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQS 120
+K GNW + + + + + ++ CL ++ ++ + T VD ++ E ++
Sbjct 1726 KKSGNWADVFTACEQALQMEPTSVQRHSDVLNCLLNMCHHQTMVTHVDGLISRVPEYKKT 1785
Query 121 RMARLAVQASWSLNKWDKMEEF 142
+ VQA+W L KWD M+E+
Sbjct 1786 WCTQ-GVQAAWRLGKWDLMDEY 1806
> cpv:cgd5_260 hypothetical protein
Length=825
Score = 32.3 bits (72), Expect = 0.45, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 5/60 (8%)
Query 4 KALHYKEKEFLEG---QTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWF 60
K LHY +E L+G +L ++ I L Y +L YSLKN+ +D + EK F
Sbjct 502 KILHYSNREVLKGFILNYPQILHCILLI--VLTNYSEYYSILNYSLKNQTNDFIISEKLF 559
> eco:b4109 yjdA, ECK4102, JW4070; mutational suppressor of YhjH
motility mutation, function unknown; related to Dynamin GTPase
Length=742
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 28 INKKLGQEQAAYGVLQYSLKNRISDVEVREKWFEKMGNWELAYLSYKF-KHEYKSDDIEA 86
+N++L + A VL Y+ ISD EVRE + L L KF + + SDD +
Sbjct 252 LNQQLARASAVLAVLDYTQLKSISDEEVREAILAVGQSVPLYVLVNKFDQQDRNSDDADQ 311
Query 87 I 87
+
Sbjct 312 V 312
> dre:572344 nuclear mitotic apparatus protein 1-like
Length=2099
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 5/62 (8%)
Query 11 KEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRIS-----DVEVREKWFEKMGN 65
+E LEG +++ +I ++ + +A LQY +K+++S + ++R + EK+GN
Sbjct 738 REELEGIVSELQAKIIEVSSIASEREACVSSLQYEMKDQLSKAKQCEADLRNEMKEKVGN 797
Query 66 WE 67
+
Sbjct 798 LQ 799
> ath:AT3G30230 myosin heavy chain-related
Length=527
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 30/119 (25%), Positives = 52/119 (43%), Gaps = 21/119 (17%)
Query 44 YSLKNRISDVEVREKWFEKMGNWELAYLSYKFKHEYKSDDIEAILG----------QSRC 93
Y+L N+ + +E + K K+ + ELA + K ++E K +E I G +S
Sbjct 367 YTLNNQFTKLEAKYKAITKLRDAELAKSAAKARNEVKGRGMELIQGAILFIQTEQARSEL 426
Query 94 LDHLGEYERLSTLVDET----FAQQDEQGQ------SRMARLAVQASWSLNKWDKMEEF 142
+ E+E L+D+ F+++ E+ + RLA + S N EEF
Sbjct 427 ESDIKEHESNLLLLDQIHKDDFSEEQERSDLKAVLYEKRIRLAALPASSFNP-QHFEEF 484
> pfa:MAL7P1.167 conserved Plasmodium protein, unknown function
Length=2773
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 25/112 (22%), Positives = 49/112 (43%), Gaps = 2/112 (1%)
Query 8 YKEKEFLEGQTTDVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWFEKMGNWE 67
+KE+E Q ++ +I + K E +Q + KN I D + E +K+ N+E
Sbjct 2268 FKEQEKNYTQNEQIIHDIIESSNKFNNENII--PIQENTKNYIDDDKYIETLDDKINNFE 2325
Query 68 LAYLSYKFKHEYKSDDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQ 119
+ K ++Y ++ +D + YE+ ST + + ++Q Q
Sbjct 2326 YIDNTIKLPNQYVPVKTSLSNNTTKNVDEINTYEKNSTFSNNNDMETNDQKQ 2377
> dre:678547 MGC136845, baat1; zgc:136845
Length=822
Score = 28.9 bits (63), Expect = 6.0, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 0/74 (0%)
Query 20 DVLGALITINKKLGQEQAAYGVLQYSLKNRISDVEVREKWFEKMGNWELAYLSYKFKHEY 79
D LGAL L + +GVL LK+ S V +K F+ + W S+ ++
Sbjct 426 DTLGALSVYEDNLDLRKDIFGVLLDYLKSPDSHATVLKKTFQAVLRWIGVCASFPDLLQF 485
Query 80 KSDDIEAILGQSRC 93
S+D+ +L + C
Sbjct 486 ISNDLFPVLEKRMC 499
> ath:AT2G11010 hypothetical protein
Length=693
Score = 28.1 bits (61), Expect = 8.8, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 44 YSLKNRISDVEVREKWFEKMGNWELAYLSYKFKHEYKSDDIEAILG 89
Y+L N+ ++++ + K K+ + ELA + K + E K IE I G
Sbjct 444 YTLNNQFTELKAKYKAIAKLRDAELAKSALKARKEVKGRRIELIQG 489
> eco:b2787 gudD, ECK2781, JW2758, ygcX; (D)-glucarate dehydratase
1 (EC:4.2.1.40); K01706 glucarate dehydratase [EC:4.2.1.40]
Length=446
Score = 28.1 bits (61), Expect = 9.8, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 5/38 (13%)
Query 82 DDIEAILGQSRCLDHLGEYERLSTLVDETFAQQDEQGQ 119
D I ++G++ LGEY+ + TLV TFA +D G+
Sbjct 67 DAIPLVVGKT-----LGEYKNVLTLVRNTFADRDAGGR 99
Lambda K H
0.315 0.130 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40