bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1577_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
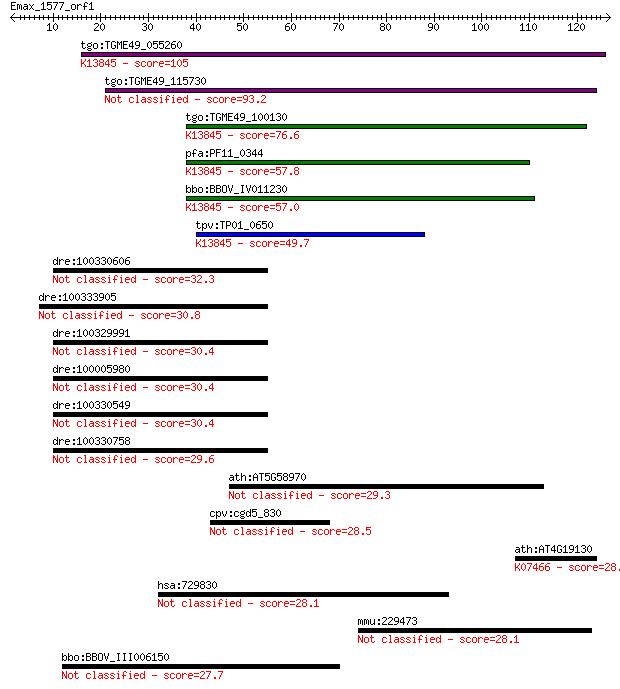
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 105 4e-23
tgo:TGME49_115730 apical membrane antigen, putative 93.2 2e-19
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 76.6 2e-14
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 57.8 8e-09
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 57.0 1e-08
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 49.7 2e-06
dre:100330606 RETRotransposon-like family member (retr-1)-like 32.3 0.41
dre:100333905 RETRotransposon-like family member (retr-1)-like 30.8 1.2
dre:100329991 LReO_3-like 30.4 1.3
dre:100005980 LReO_3-like 30.4 1.5
dre:100330549 RETRotransposon-like family member (retr-1)-like 30.4 1.5
dre:100330758 RETRotransposon-like family member (retr-1)-like 29.6 2.5
ath:AT5G58970 ATUCP2; ATUCP2 (UNCOUPLING PROTEIN 2); oxidative... 29.3 3.1
cpv:cgd5_830 membrane associated protein with a transmembrane ... 28.5 5.3
ath:AT4G19130 DNA binding / nucleic acid binding / zinc ion bi... 28.5 6.2
hsa:729830 FAM160A1, FLJ43373; family with sequence similarity... 28.1 6.6
mmu:229473 D930015E06Rik, Kiaa0922, mKIAA0922; RIKEN cDNA D930... 28.1 6.7
bbo:BBOV_III006150 17.m07545; ribosomal protein S9 27.7 8.6
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 105 bits (261), Expect = 4e-23, Method: Composition-based stats.
Identities = 56/111 (50%), Positives = 74/111 (66%), Gaps = 4/111 (3%)
Query 16 RRRRQPTDAALHQPNDNPFLVP-PLSDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREP 74
R R T +A + NPF + FM+RFN+ SGIYVDLG DKEVDG REP
Sbjct 56 RSRESQTLSA--STSGNPFQANVEMKTFMERFNLTHHHQSGIYVDLGQDKEVDGTLYREP 113
Query 75 SGLCPVFGKTIVLYQPQNNPNYKNDFLDDIPTKQQSDAVGHPLPGGFNNSF 125
+GLCP++GK I L QP + P Y+N+FL+D+PT+++ G+PLPGGFN +F
Sbjct 114 AGLCPIWGKHIELQQP-DRPPYRNNFLEDVPTEKEYKQSGNPLPGGFNLNF 163
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 57/118 (48%), Positives = 65/118 (55%), Gaps = 24/118 (20%)
Query 21 PTDAALHQPND--------------NPF-LVPPLSDFMDRFNIPKVLGSGIYVDLGGDKE 65
P DA L PN NP+ +DFM RFNIP+V GSGI+VDLG D E
Sbjct 73 PLDATLSAPNSFGEQEARSVEAVKQNPWATTTAFADFMKRFNIPQVHGSGIFVDLGRDTE 132
Query 66 VDGRTCREPSGLCPVFGKTIVLYQPQNNPNYKNDFLDDIPTKQQSDAVGHPLPGGFNN 123
RE G CPVFGK I ++QP Y N+FLDD PT +DA PLPGGFNN
Sbjct 133 ----GYREVGGKCPVFGKAIQMHQP---AEYSNNFLDDAPT--SNDASKKPLPGGFNN 181
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/85 (44%), Positives = 49/85 (57%), Gaps = 2/85 (2%)
Query 38 PLSDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREPSGLCPVFGKTIVLYQPQNNPN-Y 96
P + ++R+N+P V GSG+YVDLG K + + REP G CP +GK I YQP NP +
Sbjct 56 PWAKILERYNVPLVHGSGVYVDLGNTKILSKKKYREPGGKCPNYGKYIKTYQPTTNPEIW 115
Query 97 KNDFLDDIPTKQQSDAVGHPLPGGF 121
NDFL +P PL GGF
Sbjct 116 PNDFLKPVPYANTPQDT-MPLGGGF 139
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 32/72 (44%), Positives = 42/72 (58%), Gaps = 6/72 (8%)
Query 38 PLSDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREPSGLCPVFGKTIVLYQPQNNPNYK 97
P +++M +++I +V GSGI VDLG D EV G R PSG CPVFGK I++ N
Sbjct 109 PWTEYMAKYDIEEVHGSGIRVDLGEDAEVAGTQYRLPSGKCPVFGKGIII------ENSN 162
Query 98 NDFLDDIPTKQQ 109
FL + T Q
Sbjct 163 TTFLTPVATGNQ 174
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 41/73 (56%), Gaps = 7/73 (9%)
Query 38 PLSDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREPSGLCPVFGKTIVLYQPQNNPNYK 97
P +M +F+IP+ GSGIYVDLGG + V ++ R P G CPV GK I L
Sbjct 96 PWIKYMQKFDIPRNHGSGIYVDLGGYESVGSKSYRMPVGKCPVVGKIIDL-------GNG 148
Query 98 NDFLDDIPTKQQS 110
DFLD I ++ S
Sbjct 149 ADFLDPISSEDPS 161
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 25/48 (52%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 40 SDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREPSGLCPVFGKTIVL 87
++FM +F+I KV GSG+YVDLG V R P G CPV GK I+L
Sbjct 251 TEFMAKFDIAKVHGSGVYVDLGESATVGIYDYRMPIGKCPVVGKAIIL 298
> dre:100330606 RETRotransposon-like family member (retr-1)-like
Length=722
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 10 LVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
L+ IMC+ R P L L L+DFM F IPKV+ S
Sbjct 69 LLTIMCQTTRYPAAYPLRSITTKSIL-KALTDFMSIFGIPKVIQS 112
> dre:100333905 RETRotransposon-like family member (retr-1)-like
Length=761
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Query 7 TKQLVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
+K L+ +MC+ R P AL +V LS F+ F IPKV+ S
Sbjct 347 SKFLLTVMCQSTRYPAAYALRNITTRS-VVKALSQFISIFGIPKVIQS 393
> dre:100329991 LReO_3-like
Length=1120
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 10 LVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
L+ +MC+ R PT L L L++FM F IPK + S
Sbjct 386 LLTVMCQSTRYPTAYPLRSITTKSIL-KALTNFMSIFGIPKTIQS 429
> dre:100005980 LReO_3-like
Length=1250
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 10 LVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
L+ +MC+ R PT L L L++FM F IPK + S
Sbjct 502 LLTVMCQSTRYPTAYPLRSITTKSIL-KALTNFMSIFGIPKTIQS 545
> dre:100330549 RETRotransposon-like family member (retr-1)-like
Length=1039
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 10 LVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
L+ +MC+ R PT L L L++FM F IPK + S
Sbjct 388 LLTVMCQSTRYPTAYPLRSITTKSIL-KALTNFMSIFGIPKTIQS 431
> dre:100330758 RETRotransposon-like family member (retr-1)-like
Length=549
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 10 LVQIMCRRRRQPTDAALHQPNDNPFLVPPLSDFMDRFNIPKVLGS 54
L+ IMC+ P +L L L+DFM F IPKV+ S
Sbjct 87 LLTIMCQTTHYPAAYSLRSITTKSIL-KALTDFMSIFGIPKVIQS 130
> ath:AT5G58970 ATUCP2; ATUCP2 (UNCOUPLING PROTEIN 2); oxidative
phosphorylation uncoupler; K15103 solute carrier family 25
(mitochondrial uncoupling protein), member 8/9
Length=305
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 4/70 (5%)
Query 47 NIPKVLGS-GIYVDLGGDKEVDGRTCREPSGL---CPVFGKTIVLYQPQNNPNYKNDFLD 102
N+PK GS G + ++ + G +GL C G I LY+P +DF+
Sbjct 53 NLPKYRGSIGTLATIAREEGISGLWKGVIAGLHRQCIYGGLRIGLYEPVKTLLVGSDFIG 112
Query 103 DIPTKQQSDA 112
DIP Q+ A
Sbjct 113 DIPLYQKILA 122
> cpv:cgd5_830 membrane associated protein with a transmembrane
domain near C-terminus
Length=901
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Query 43 MDRFNIPKVLGS-GIYVDLGGDKEVD 67
+D+FN P+VLG +VD GDK D
Sbjct 54 LDKFNFPEVLGKIKSFVDFNGDKFTD 79
> ath:AT4G19130 DNA binding / nucleic acid binding / zinc ion
binding; K07466 replication factor A1
Length=784
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 11/17 (64%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 107 KQQSDAVGHPLPGGFNN 123
KQ SD +GHP+PGG +N
Sbjct 106 KQISDIIGHPVPGGKHN 122
> hsa:729830 FAM160A1, FLJ43373; family with sequence similarity
160, member A1
Length=1040
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 34/61 (55%), Gaps = 7/61 (11%)
Query 32 NPFLVPPLSDFMDRFNIPKVLGSGIYVDLGGDKEVDGRTCREPSGLCPVFGKTIVLYQPQ 91
N FLVP L+ + + + +V+ + Y+DL R+ EP+ L +F + I+L+Q +
Sbjct 313 NGFLVPVLAPALHKVTVEEVMTTTAYLDLF------LRSISEPA-LLEIFLRFILLHQHE 365
Query 92 N 92
N
Sbjct 366 N 366
> mmu:229473 D930015E06Rik, Kiaa0922, mKIAA0922; RIKEN cDNA D930015E06
gene
Length=1597
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Query 74 PSGLCPVFGKTIVLYQPQNNPNYKNDFLDDIPTKQQSDAVGHPLPGGFN 122
PS CP+ + P+ N NY N F +K Q+D +GH P +N
Sbjct 1444 PSVYCPLELNDYNAF-PEENMNYTNGF--PCSSKVQTDFIGHSTPSTWN 1489
> bbo:BBOV_III006150 17.m07545; ribosomal protein S9
Length=683
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 24/61 (39%), Gaps = 3/61 (4%)
Query 12 QIMCRRRRQPTDAA---LHQPNDNPFLVPPLSDFMDRFNIPKVLGSGIYVDLGGDKEVDG 68
Q+ RRRR P + DN L MD FNIPK VD+ + G
Sbjct 191 QLKSRRRRLPQSFYGDDTYSSVDNEKLPEGCKSLMDFFNIPKNFDFKALVDVSTQSSLSG 250
Query 69 R 69
R
Sbjct 251 R 251
Lambda K H
0.320 0.142 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40