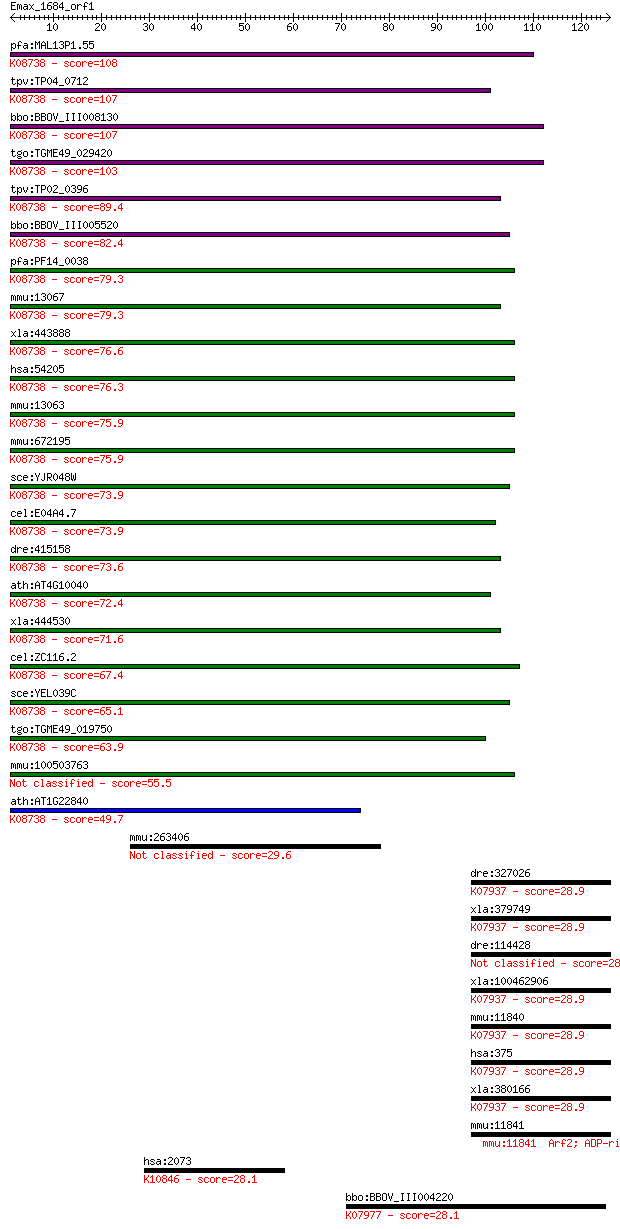
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1684_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
pfa:MAL13P1.55 cytochrome c2 precursor,putative; K08738 cytoch... 108 6e-24
tpv:TP04_0712 cytochrome c; K08738 cytochrome c 107 9e-24
bbo:BBOV_III008130 17.m07711; cytochrome c family protein; K08... 107 9e-24
tgo:TGME49_029420 cytochrome c, putative ; K08738 cytochrome c 103 2e-22
tpv:TP02_0396 cytochrome c; K08738 cytochrome c 89.4 3e-18
bbo:BBOV_III005520 17.m07493; cytochrome c; K08738 cytochrome c 82.4 3e-16
pfa:PF14_0038 cytochrome c, putative; K08738 cytochrome c 79.3 3e-15
mmu:13067 Cyct, T-Cc; cytochrome c, testis; K08738 cytochrome c 79.3 3e-15
xla:443888 cyct-a, MGC80124, cycs-a; cytochrome c, testis; K08... 76.6 2e-14
hsa:54205 CYCS, CYC, HCS, THC4; cytochrome c, somatic; K08738 ... 76.3 2e-14
mmu:13063 Cycs; cytochrome c, somatic; K08738 cytochrome c 75.9 3e-14
mmu:672195 Gm10053; predicted gene 10053; K08738 cytochrome c 75.9 3e-14
sce:YJR048W CYC1; Cyc1p; K08738 cytochrome c 73.9 1e-13
cel:E04A4.7 cyc-2.1; CYtochrome C family member (cyc-2.1); K08... 73.9 1e-13
dre:415158 cycsb, cyc, wu:fk52b01, zgc:86706; cytochrome c, so... 73.6 2e-13
ath:AT4G10040 CYTC-2; CYTC-2 (cytochrome c-2); electron carrie... 72.4 3e-13
xla:444530 cycs, cyct; cytochrome c, somatic; K08738 cytochrome c 71.6 6e-13
cel:ZC116.2 cyc-2.2; CYtochrome C family member (cyc-2.2); K08... 67.4 1e-11
sce:YEL039C CYC7; Cyc7p; K08738 cytochrome c 65.1 5e-11
tgo:TGME49_019750 cytochrome c, putative ; K08738 cytochrome c 63.9 1e-10
mmu:100503763 cytochrome c, somatic-like 55.5 4e-08
ath:AT1G22840 CYTC-1; CYTC-1 (CYTOCHROME C-1); electron carrie... 49.7 2e-06
mmu:263406 Plekhg3, BC030417, MGC40768; pleckstrin homology do... 29.6 2.2
dre:327026 arf1l, fa19d09, wu:fa19d09, wu:fb19d05; ADP-ribosyl... 28.9 4.2
xla:379749 arf1, MGC52628, arf2; ADP-ribosylation factor 1; K0... 28.9 4.2
dre:114428 arf1, wu:fb33e02, wu:fb78h01; ADP-ribosylation fact... 28.9 4.4
xla:100462906 arf-1; arf-1 protein; K07937 ADP-ribosylation fa... 28.9 4.6
mmu:11840 Arf1; ADP-ribosylation factor 1; K07937 ADP-ribosyla... 28.9 4.6
hsa:375 ARF1; ADP-ribosylation factor 1; K07937 ADP-ribosylati... 28.9 4.6
xla:380166 arf5, MGC52573, arf-1, arf1; ADP-ribosylation facto... 28.9 4.8
mmu:11841 Arf2; ADP-ribosylation factor 2; K06883 28.5 5.0
hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair... 28.1 6.5
bbo:BBOV_III004220 17.m07380; ADP-ribosylation factor family p... 28.1 7.6
> pfa:MAL13P1.55 cytochrome c2 precursor,putative; K08738 cytochrome
c
Length=159
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 50/110 (45%), Positives = 68/110 (61%), Gaps = 1/110 (0%)
Query 1 LFKKHCAQCHVIEKDQS-VKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVW 59
LFKKHC QCH I D S +G T GP+L+ VY RTAG S +S + SGI+W
Sbjct 33 LFKKHCKQCHSIAPDNSQTNSGFTSWGPTLFNVYNRTAGMSKGNSPFQTSPDLYTSGIIW 92
Query 60 TDKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACEDKTEG 109
D L++Y+KNP+QF + + MNF+GL + Q+RVD++ Y+K D G
Sbjct 93 NDVNLLKYMKNPQQFVESHIGMNFKGLSNLQERVDIVHYLKTLTYDDPYG 142
> tpv:TP04_0712 cytochrome c; K08738 cytochrome c
Length=145
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 47/100 (47%), Positives = 66/100 (66%), Gaps = 0/100 (0%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFKKHC QCH + D G + +GP+L+ VYGRT+G L +H++K SGIVW
Sbjct 27 LFKKHCKQCHSMRPDNRQTGGFSSIGPTLFNVYGRTSGIQNLGGLNLMTHSMKSSGIVWD 86
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMK 100
D LMRY+KNP+ F + + MNF GL +QDRVD++ +++
Sbjct 87 DANLMRYMKNPQLFVNNKIGMNFAGLPRFQDRVDIVHFLR 126
> bbo:BBOV_III008130 17.m07711; cytochrome c family protein; K08738
cytochrome c
Length=146
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 47/111 (42%), Positives = 70/111 (63%), Gaps = 0/111 (0%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFKKHC QCH + D +G +GP+L+ VY RTAG+ S+ + ++ +GIVWT
Sbjct 27 LFKKHCQQCHSMRPDNRQTSGFATVGPTLFNVYCRTAGASGHDSVKGITDNLQNAGIVWT 86
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACEDKTEGLK 111
D LMRY+KNP ++ + MNF GL ++QDRVD++ +++ + T G K
Sbjct 87 DANLMRYMKNPERYVNAVVGMNFSGLPNFQDRVDIVHFLRDLTPEGTVGKK 137
> tgo:TGME49_029420 cytochrome c, putative ; K08738 cytochrome
c
Length=163
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 53/112 (47%), Positives = 69/112 (61%), Gaps = 1/112 (0%)
Query 1 LFKKHCAQCHVIEKD-QSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVW 59
LFKKHCAQCH I D + + G T GP+L+ VY RTAG +S S I +SG+VW
Sbjct 41 LFKKHCAQCHSIFPDGRHLIAGNTSWGPTLWNVYMRTAGVEKDSSCSPISSHILDSGVVW 100
Query 60 TDKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACEDKTEGLK 111
D LMRY+KNP+ F + MNF G+ ++QDRVD+I Y+K + G K
Sbjct 101 NDANLMRYMKNPKMFINGVVGMNFFGIANFQDRVDIIHYLKTLTWNHPNGKK 152
> tpv:TP02_0396 cytochrome c; K08738 cytochrome c
Length=115
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 52/102 (50%), Positives = 59/102 (57%), Gaps = 10/102 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFK CAQCH I K SVK G P+LYG YGR +G+ S S A K SGIVW+
Sbjct 21 LFKSKCAQCHTINKGGSVKQG-----PNLYGFYGRKSGA----SDYAYSDANKNSGIVWS 71
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAA 102
DK L YL NP+Q+ M F GL QDR DLI Y+K A
Sbjct 72 DKHLFVYLVNPKQYIPG-TKMVFAGLKKEQDRADLIAYLKEA 112
> bbo:BBOV_III005520 17.m07493; cytochrome c; K08738 cytochrome
c
Length=115
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 58/104 (55%), Gaps = 10/104 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFK CAQCH K G T GP+LYG++GR +GS + A K SGIVW+
Sbjct 21 LFKSKCAQCHTTNKG-----GATKQGPNLYGLFGRASGSADYAY----TDANKSSGIVWS 71
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACE 104
DK L YL NP+++ M F GL QDR D+I Y+K A +
Sbjct 72 DKHLFVYLVNPKEYIPG-TKMIFAGLKKEQDRADIIAYLKEATK 114
> pfa:PF14_0038 cytochrome c, putative; K08738 cytochrome c
Length=115
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/105 (42%), Positives = 58/105 (55%), Gaps = 10/105 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFK CAQCH I K +VK G P+L+G YGR +G S S A K SGI+W+
Sbjct 21 LFKAKCAQCHTINKGGAVKQG-----PNLHGFYGRKSGD----SDFPYSDANKNSGIIWS 71
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
DK L YL NP+ + M F G+ ++R DLI Y+K A +
Sbjct 72 DKHLFEYLLNPKLYIPG-TKMIFAGIKKEKERADLIEYLKKASSE 115
> mmu:13067 Cyct, T-Cc; cytochrome c, testis; K08738 cytochrome
c
Length=105
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 61/102 (59%), Gaps = 9/102 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH +EK KTG P+L+G++GR G P S D++ K G++W+
Sbjct 10 IFVQKCAQCHTVEKGGKHKTG-----PNLWGLFGRKTGQAPGFSYTDAN---KNKGVIWS 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAA 102
++TLM YL+NP+++ M F G+ +R DLI Y+K A
Sbjct 62 EETLMEYLENPKKYIPG-TKMIFAGIKKKSEREDLIKYLKQA 102
> xla:443888 cyct-a, MGC80124, cycs-a; cytochrome c, testis; K08738
cytochrome c
Length=105
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 60/105 (57%), Gaps = 9/105 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH +EK KTG P+L+G++GR G P S D++ K GIVW
Sbjct 10 IFIQKCAQCHTVEKTGKHKTG-----PNLWGLFGRKTGQAPGFSYTDAN---KSKGIVWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
+ TL YL+NP+++ M F G+ +R+DLI Y+K + +
Sbjct 62 EDTLFEYLENPKKYIPG-TKMIFAGIKKKNERLDLIAYLKKSTSE 105
> hsa:54205 CYCS, CYC, HCS, THC4; cytochrome c, somatic; K08738
cytochrome c
Length=105
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 59/105 (56%), Gaps = 9/105 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F C+QCH +EK KTG P+L+G++GR G P S + A K GI+W
Sbjct 10 IFIMKCSQCHTVEKGGKHKTG-----PNLHGLFGRKTGQAPGYSY---TAANKNKGIIWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
+ TLM YL+NP+++ M F G+ ++R DLI Y+K A +
Sbjct 62 EDTLMEYLENPKKYIPG-TKMIFVGIKKKEERADLIAYLKKATNE 105
> mmu:13063 Cycs; cytochrome c, somatic; K08738 cytochrome c
Length=105
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 58/105 (55%), Gaps = 9/105 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH +EK KTG P+L+G++GR G S D++ K GI W
Sbjct 10 IFVQKCAQCHTVEKGGKHKTG-----PNLHGLFGRKTGQAAGFSYTDAN---KNKGITWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
+ TLM YL+NP+++ M F G+ +R DLI Y+K A +
Sbjct 62 EDTLMEYLENPKKYIP-GTKMIFAGIKKKGERADLIAYLKKATNE 105
> mmu:672195 Gm10053; predicted gene 10053; K08738 cytochrome
c
Length=105
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/105 (38%), Positives = 58/105 (55%), Gaps = 9/105 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH +EK KTG P+L+G++GR G S D++ K GI W
Sbjct 10 IFVQKCAQCHTVEKGGKHKTG-----PNLHGLFGRKTGQAAGFSYTDAN---KNKGITWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
+ TLM YL+NP+++ M F G+ +R DLI Y+K A +
Sbjct 62 EDTLMEYLENPKKYIP-GTKMIFAGIKKKGERADLIAYLKKATNE 105
> sce:YJR048W CYC1; Cyc1p; K08738 cytochrome c
Length=109
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 60/104 (57%), Gaps = 9/104 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFK C QCH +EK K +GP+L+G++GR +G S D++ IK++ ++W
Sbjct 15 LFKTRCLQCHTVEKGGPHK-----VGPNLHGIFGRHSGQAEGYSYTDAN--IKKN-VLWD 66
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACE 104
+ + YL NP+++ M F GL +DR DLI Y+K ACE
Sbjct 67 ENNMSEYLTNPKKYIP-GTKMAFGGLKKEKDRNDLITYLKKACE 109
> cel:E04A4.7 cyc-2.1; CYtochrome C family member (cyc-2.1); K08738
cytochrome c
Length=111
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 58/101 (57%), Gaps = 10/101 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
++K+ C QCHV++ + T GP+L+GV GRT+G+ S D S A K G+VWT
Sbjct 15 VYKQRCLQCHVVD------STATKTGPTLHGVIGRTSGT---VSGFDYSAANKNKGVVWT 65
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKA 101
+TL YL NP+++ M F GL +R DLI Y++
Sbjct 66 RETLFEYLLNPKKYIPG-TKMVFAGLKKADERADLIKYIEV 105
> dre:415158 cycsb, cyc, wu:fk52b01, zgc:86706; cytochrome c,
somatic b; K08738 cytochrome c
Length=104
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 58/102 (56%), Gaps = 9/102 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH +E K +GP+L+G++GR G S D++ K GIVW
Sbjct 10 VFVQKCAQCHTVENGGKHK-----VGPNLWGLFGRKTGQAEGFSYTDAN---KSKGIVWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAA 102
+ TLM YL+NP+++ M F G+ +R DLI Y+K+A
Sbjct 62 EDTLMEYLENPKKYIPG-TKMIFAGIKKKGERADLIAYLKSA 102
> ath:AT4G10040 CYTC-2; CYTC-2 (cytochrome c-2); electron carrier/
heme binding / iron ion binding; K08738 cytochrome c
Length=112
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 41/100 (41%), Positives = 55/100 (55%), Gaps = 9/100 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F+ CAQCH +EK K GP+L G++GR +G+ P S S A K + W
Sbjct 18 IFRTKCAQCHTVEKGAGHKQ-----GPNLNGLFGRQSGTTPGYSY---SAANKSMAVNWE 69
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMK 100
+KTL YL NP+++ M F GL QDR DLI Y+K
Sbjct 70 EKTLYDYLLNPKKYIPG-TKMVFPGLKKPQDRADLIAYLK 108
> xla:444530 cycs, cyct; cytochrome c, somatic; K08738 cytochrome
c
Length=105
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 58/102 (56%), Gaps = 9/102 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + C+QCH +EK KTG P+L+G++GR G S D++ K GIVW
Sbjct 10 VFVQKCSQCHTVEKGGKHKTG-----PNLHGLFGRKTGQAEGFSYTDAN---KNKGIVWD 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAA 102
+ TLM YL+NP+++ M F G+ +R DLI Y+K +
Sbjct 62 EDTLMVYLENPKKYIP-GTKMIFAGIKKKGERQDLIAYLKQS 102
> cel:ZC116.2 cyc-2.2; CYtochrome C family member (cyc-2.2); K08738
cytochrome c
Length=123
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/106 (35%), Positives = 55/106 (51%), Gaps = 10/106 (9%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+FK+ C QCHV+ Q T GP+L GV GR +G + + D S A K G+VW
Sbjct 25 IFKQRCEQCHVVNSLQ------TKTGPTLNGVIGRQSG---QVAGFDYSAANKNKGVVWD 75
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACEDK 106
+TL YL +P+++ M F GL +R DLI +++ K
Sbjct 76 RQTLFDYLADPKKYIPG-TKMVFAGLKKADERADLIKFIEVEAAKK 120
> sce:YEL039C CYC7; Cyc7p; K08738 cytochrome c
Length=113
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 54/104 (51%), Gaps = 9/104 (8%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
LFK C QCH IE+ G +GP+L+G++GR +G S D++ + W
Sbjct 19 LFKTRCQQCHTIEEG-----GPNKVGPNLHGIFGRHSGQVKGYSYTDANI---NKNVKWD 70
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACE 104
+ ++ YL NP+++ M F GL +DR DLI YM A +
Sbjct 71 EDSMSEYLTNPKKYIPG-TKMAFAGLKKEKDRNDLITYMTKAAK 113
> tgo:TGME49_019750 cytochrome c, putative ; K08738 cytochrome
c
Length=115
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 52/99 (52%), Gaps = 10/99 (10%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+FK C+QCH I + K G P+L+G GR +G + S A K SGI+W+
Sbjct 21 VFKAKCSQCHTINAGGAAKAG-----PNLHGFIGRESGK----ADFPYSDANKTSGIIWS 71
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYM 99
+K L YL NP+ + M F G+ ++R DLI Y+
Sbjct 72 EKHLWEYLLNPKTYIPG-TKMVFAGIKKEKERADLIAYL 109
> mmu:100503763 cytochrome c, somatic-like
Length=106
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 50/105 (47%), Gaps = 8/105 (7%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F + CAQCH + K KT GP+L G++G+ G S D+S K GI W
Sbjct 10 IFVQRCAQCHTVRKGGKHKT-----GPNLQGLFGQKTGQAAGFSYTDAS---KNRGITWG 61
Query 61 DKTLMRYLKNPRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACED 105
+ TL+ + Q M F G+ ++ DLI Y+K A +
Sbjct 62 EDTLLDGVFRESQKYIPGTKMIFAGIKKTGEKKDLIAYLKKAANE 106
> ath:AT1G22840 CYTC-1; CYTC-1 (CYTOCHROME C-1); electron carrier/
heme binding / iron ion binding; K08738 cytochrome c
Length=102
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 39/73 (53%), Gaps = 8/73 (10%)
Query 1 LFKKHCAQCHVIEKDQSVKTGCTLLGPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWT 60
+F+ CAQCH +E K GP+L G++GR +G+ T+ S A K + W
Sbjct 18 IFRTKCAQCHTVEAGAGHKQ-----GPNLNGLFGRQSGT---TAGYSYSAANKNKAVEWE 69
Query 61 DKTLMRYLKNPRQ 73
+K L YL NP++
Sbjct 70 EKALYDYLLNPKK 82
> mmu:263406 Plekhg3, BC030417, MGC40768; pleckstrin homology
domain containing, family G (with RhoGef domain) member 3
Length=1341
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 26 GPSLYGVYGRTAGSGPRTSLLDSSHAIKESGIVWTDKTLMRYLKNPRQFTQH 77
GPS+ G +AGSG T SS KES + D+ L+ +KN + +H
Sbjct 799 GPSVNGTESPSAGSGCPTEQDRSSCKKKESALSTRDRQLLDKIKNYYENAEH 850
> dre:327026 arf1l, fa19d09, wu:fa19d09, wu:fb19d05; ADP-ribosylation
factor 1 like; K07937 ADP-ribosylation factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLKNQK 181
> xla:379749 arf1, MGC52628, arf2; ADP-ribosylation factor 1;
K07937 ADP-ribosylation factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLKNQK 181
> dre:114428 arf1, wu:fb33e02, wu:fb78h01; ADP-ribosylation factor
1
Length=180
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 152 WYIQATCATSGDGLYEGLDWLSNQLKNQK 180
> xla:100462906 arf-1; arf-1 protein; K07937 ADP-ribosylation
factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLRNQK 181
> mmu:11840 Arf1; ADP-ribosylation factor 1; K07937 ADP-ribosylation
factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLRNQK 181
> hsa:375 ARF1; ADP-ribosylation factor 1; K07937 ADP-ribosylation
factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLRNQK 181
> xla:380166 arf5, MGC52573, arf-1, arf1; ADP-ribosylation factor
5; K07937 ADP-ribosylation factor 1
Length=181
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLRNQK 181
> mmu:11841 Arf2; ADP-ribosylation factor 2; K06883
Length=181
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 97 WYMKAACEDKTEGLKEGLSSIRTRLQQQQ 125
WY++A C +GL EGL + +L+ Q+
Sbjct 153 WYIQATCATSGDGLYEGLDWLSNQLKNQK 181
> hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair
cross-complementing rodent repair deficiency, complementation
group 5; K10846 DNA excision repair protein ERCC-5
Length=1186
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 29 LYGVYGRTAGSGPRTSLLDSSHAIKESGI 57
L V RTA P+TS DS +++KE+ +
Sbjct 1110 LMNVQRRTAAKEPKTSASDSQNSVKEAPV 1138
> bbo:BBOV_III004220 17.m07380; ADP-ribosylation factor family
protein; K07977 Arf/Sar family, other
Length=180
Score = 28.1 bits (61), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 27/54 (50%), Gaps = 4/54 (7%)
Query 71 PRQFTQHPLSMNFRGLDSWQDRVDLIWYMKAACEDKTEGLKEGLSSIRTRLQQQ 124
P LS N GL+ + +R WY++AAC + GL EG+ + L+ +
Sbjct 131 PNAIPATELS-NELGLNQFGNRT---WYIQAACATQGTGLYEGMDWLTNALKHK 180
Lambda K H
0.319 0.133 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40