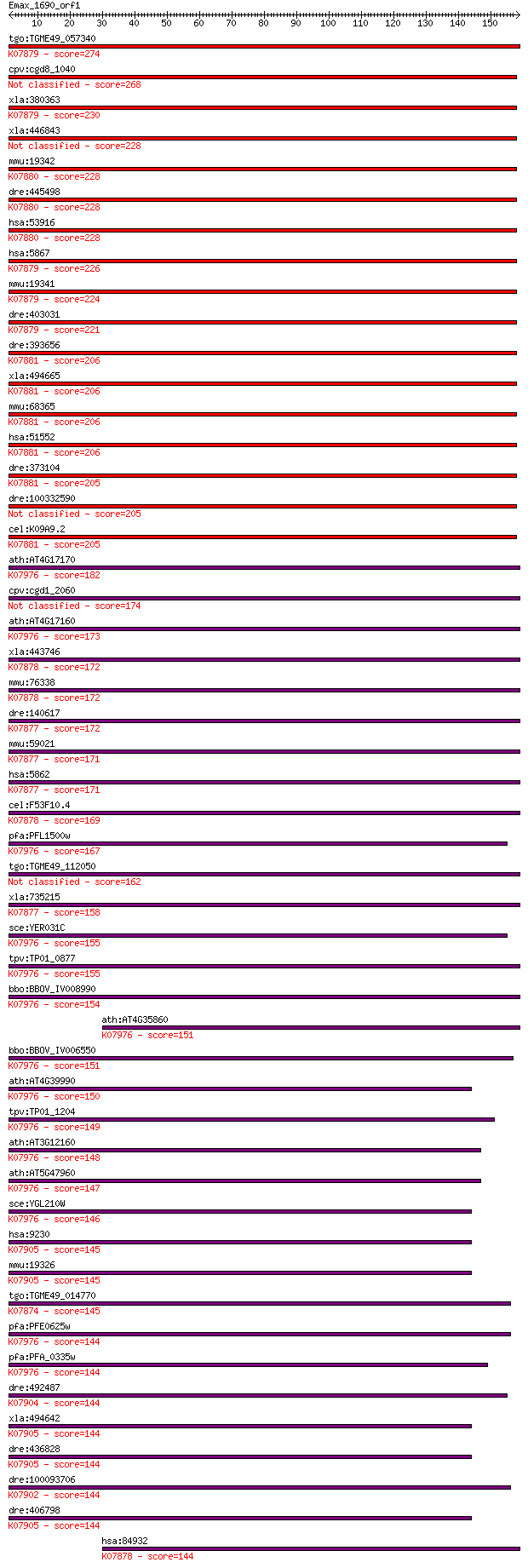
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1690_orf1
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_057340 Ras family domain-containing protein (EC:3.1... 274 9e-74
cpv:cgd8_1040 hypothetical protein 268 5e-72
xla:380363 rab4a, MGC52945; RAB4A, member RAS oncogene family;... 230 1e-60
xla:446843 rab4b, MGC80680, rab14; RAB4B member RAS oncogene f... 228 6e-60
mmu:19342 Rab4b, 1500031G17Rik; RAB4B, member RAS oncogene fam... 228 6e-60
dre:445498 rab4b; zgc:101015; K07880 Ras-related protein Rab-4B 228 6e-60
hsa:53916 RAB4B, FLJ78649, MGC52123; RAB4B, member RAS oncogen... 228 6e-60
hsa:5867 RAB4A, HRES-1/RAB4, RAB4; RAB4A, member RAS oncogene ... 226 3e-59
mmu:19341 Rab4a, AI848268, Rab4; RAB4A, member RAS oncogene fa... 224 1e-58
dre:403031 rab4a, fc93b03, wu:fc93b03; RAB4a, member RAS oncog... 221 8e-58
dre:393656 rab14l, MGC63643, zgc:63643; RAB14, member RAS onco... 206 3e-53
xla:494665 rab14; RAB14, member RAS oncogene family; K07881 Ra... 206 3e-53
mmu:68365 Rab14, 0610030G24Rik, 2810475J17Rik, A830021G03Rik, ... 206 3e-53
hsa:51552 RAB14, FBP, RAB-14; RAB14, member RAS oncogene famil... 206 3e-53
dre:373104 rab14, cb731, fc26g11, fj47f07, fj68a01, wu:fc26g11... 205 5e-53
dre:100332590 RAB14, member RAS oncogene family-like 205 5e-53
cel:K09A9.2 rab-14; RAB family member (rab-14); K07881 Ras-rel... 205 5e-53
ath:AT4G17170 RABB1C; RABB1C (ARABIDOPSIS RAB GTPASE HOMOLOG B... 182 3e-46
cpv:cgd1_2060 Rab2 GTPase 174 8e-44
ath:AT4G17160 ATRABB1A (ARABIDOPSIS RAB GTPASE HOMOLOG B1A); G... 173 2e-43
xla:443746 rab2b, MGC132078, MGC78967; rab2b, member RAS oncog... 172 3e-43
mmu:76338 Rab2b, 1500012D09Rik, 4930528G15Rik, A230002G14, D53... 172 3e-43
dre:140617 rab2a, cb75, fb57e06, rab2, wu:fb57e06; RAB2A, memb... 172 6e-43
mmu:59021 Rab2a, 9330148M11Rik, C80220, Rab2; RAB2A, member RA... 171 7e-43
hsa:5862 RAB2A, RAB2; RAB2A, member RAS oncogene family; K0787... 171 7e-43
cel:F53F10.4 unc-108; UNCoordinated family member (unc-108); K... 169 4e-42
pfa:PFL1500w rab2; Rab2, GTPase (EC:3.6.1.-); K07976 Rab famil... 167 9e-42
tgo:TGME49_112050 Ras family domain-containing protein (EC:3.1... 162 6e-40
xla:735215 rab2a, MGC131016, rab2; RAB2A, member RAS oncogene ... 158 8e-39
sce:YER031C YPT31, YPT8; GTPase of the Ypt/Rab family, very si... 155 5e-38
tpv:TP01_0877 hypothetical protein; K07976 Rab family, other 155 5e-38
bbo:BBOV_IV008990 23.m06418; Rab2; K07976 Rab family, other 154 1e-37
ath:AT4G35860 ATGB2; ATGB2 (GTP-BINDING 2); GTP binding; K0797... 151 1e-36
bbo:BBOV_IV006550 23.m06035; ras-related protein Rab11A; K0797... 151 1e-36
ath:AT4G39990 RABA4B; RABA4B (RAB GTPASE HOMOLOG A4B); GTP bin... 150 2e-36
tpv:TP01_1204 hypothetical protein; K07976 Rab family, other 149 3e-36
ath:AT3G12160 RABA4D; RABA4D (RAB GTPASE HOMOLOG A4D); GTP bin... 148 9e-36
ath:AT5G47960 ATRABA4C; GTP binding; K07976 Rab family, other 147 1e-35
sce:YGL210W YPT32; GTPase of the Ypt/Rab family, very similar ... 146 3e-35
hsa:9230 RAB11B, H-YPT3, MGC133246; RAB11B, member RAS oncogen... 145 5e-35
mmu:19326 Rab11b, A730055L17Rik; RAB11B, member RAS oncogene f... 145 5e-35
tgo:TGME49_014770 RAS small GTpase, putative (EC:3.1.4.12); K0... 145 7e-35
pfa:PFE0625w Rab1b; Rab1b, GTPase; K07976 Rab family, other 144 9e-35
pfa:PFA_0335w Rab5c; Rab5c, GTPase (EC:3.6.5.2); K07976 Rab fa... 144 1e-34
dre:492487 rab11a, wu:fi15h09, zgc:103679; RAB11a, member RAS ... 144 1e-34
xla:494642 hypothetical LOC494642; K07905 Ras-related protein ... 144 1e-34
dre:436828 zgc:92772; K07905 Ras-related protein Rab-11B 144 1e-34
dre:100093706 rab8b, zgc:165529; RAB8B, member RAS oncogene fa... 144 1e-34
dre:406798 rab11b, wu:fb07g11, zgc:55760; RAB11B, member RAS o... 144 1e-34
hsa:84932 RAB2B, FLJ14824; RAB2B, member RAS oncogene family; ... 144 2e-34
> tgo:TGME49_057340 Ras family domain-containing protein (EC:3.1.4.12);
K07879 Ras-related protein Rab-4A
Length=214
Score = 274 bits (700), Expect = 9e-74, Method: Compositional matrix adjust.
Identities = 126/158 (79%), Positives = 148/158 (93%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+ENKF++ SHTIGVEF SK++N+ +IKLQIWDTAGQERYR+VTRSYYR
Sbjct 26 GKSCLLHQFIENKFKKGCSHTIGVEFGSKLINIAGKRIKLQIWDTAGQERYRSVTRSYYR 85
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD++NR+SYNHL+NWLADARSLARADISI VGNK+D+++KRAV+FLEASRF
Sbjct 86 GAAGALLVYDISNRDSYNHLMNWLADARSLARADISIIAVGNKVDLKEKRAVSFLEASRF 145
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
AQEND+LFLETSA TGEGV DVFV+VARLIL+KI+DGL
Sbjct 146 AQENDILFLETSAFTGEGVEDVFVKVARLILNKIEDGL 183
> cpv:cgd8_1040 hypothetical protein
Length=216
Score = 268 bits (686), Expect = 5e-72, Method: Compositional matrix adjust.
Identities = 123/157 (78%), Positives = 142/157 (90%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+ENKF++ SSHTIGVEF SK++ +IKLQIWDTAGQERYR+VTRSYYR
Sbjct 24 GKSCLLHQFIENKFKKGSSHTIGVEFGSKIITAGGKRIKLQIWDTAGQERYRSVTRSYYR 83
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ NRESYNHL+NWLADAR+LAR DISI VGNK+D++DKR VTFLEASR
Sbjct 84 GAAGALIVYDITNRESYNHLVNWLADARTLARVDISIIGVGNKLDLKDKRQVTFLEASRC 143
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEND+LFLETSALTGEGV +VFV+V RLILSKI+DG
Sbjct 144 AQENDILFLETSALTGEGVEEVFVKVTRLILSKIEDG 180
> xla:380363 rab4a, MGC52945; RAB4A, member RAS oncogene family;
K07879 Ras-related protein Rab-4A
Length=213
Score = 230 bits (587), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 105/157 (66%), Positives = 132/157 (84%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+E+KF++ S+HTIGVEF S+++N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIESKFKQDSNHTIGVEFGSRIVNVGGKSVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+A+RE+YN L NWL DAR+LA +I I + GNK D+ R VTFLEASRF
Sbjct 80 GAAGALLVYDIASRETYNALTNWLTDARTLASPNIIIILCGNKKDLDADREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR ILSKI+ G
Sbjct 140 AQENELMFLETSALTGENVEEAFLKCARTILSKIESG 176
> xla:446843 rab4b, MGC80680, rab14; RAB4B member RAS oncogene
family
Length=213
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 104/157 (66%), Positives = 132/157 (84%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+E+KF++ S+HTIGVEF S+++N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIESKFKQDSNHTIGVEFGSRIVNVASKSVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+A+RE+YN L NWL DAR+LA +I I + GNK D+ R VTFLEASRF
Sbjct 80 GAAGALLVYDIASRETYNALTNWLTDARTLASPNIIIILCGNKKDLDADREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR IL+KI+ G
Sbjct 140 AQENELMFLETSALTGENVEEAFLKCARTILNKIESG 176
> mmu:19342 Rab4b, 1500031G17Rik; RAB4B, member RAS oncogene family;
K07880 Ras-related protein Rab-4B
Length=213
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 103/157 (65%), Positives = 130/157 (82%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+ENKF++ S+HTIGVEF S+V+N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIENKFKQDSNHTIGVEFGSRVVNVGGKTVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L WL DAR+LA +I + + GNK D+ +R VTFLEASRF
Sbjct 80 GAAGALLVYDITSRETYNSLAAWLTDARTLASPNIVVILCGNKKDLDPEREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR IL+KI G
Sbjct 140 AQENELMFLETSALTGENVEEAFLKCARTILNKIDSG 176
> dre:445498 rab4b; zgc:101015; K07880 Ras-related protein Rab-4B
Length=213
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 104/157 (66%), Positives = 130/157 (82%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+ENKF++ S+HTIGVEF S+V+N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIENKFKQDSNHTIGVEFGSRVVNVGGKTVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L NWL DAR+LA +I I + GNK D+ R VTFLEASRF
Sbjct 80 GAAGALLVYDITSRETYNALTNWLTDARTLASPNIVIILCGNKKDLDADREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR I +KI+ G
Sbjct 140 AQENELMFLETSALTGENVEEAFLKCARSIPNKIESG 176
> hsa:53916 RAB4B, FLJ78649, MGC52123; RAB4B, member RAS oncogene
family; K07880 Ras-related protein Rab-4B
Length=213
Score = 228 bits (581), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 103/157 (65%), Positives = 130/157 (82%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+ENKF++ S+HTIGVEF S+V+N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIENKFKQDSNHTIGVEFGSRVVNVGGKTVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L WL DAR+LA +I + + GNK D+ +R VTFLEASRF
Sbjct 80 GAAGALLVYDITSRETYNSLAAWLTDARTLASPNIVVILCGNKKDLDPEREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR IL+KI G
Sbjct 140 AQENELMFLETSALTGENVEEAFLKCARTILNKIDSG 176
> hsa:5867 RAB4A, HRES-1/RAB4, RAB4; RAB4A, member RAS oncogene
family; K07879 Ras-related protein Rab-4A
Length=218
Score = 226 bits (575), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 105/157 (66%), Positives = 128/157 (81%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+E KF+ S+HTIGVEF SK++N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 25 GKSCLLHQFIEKKFKDDSNHTIGVEFGSKIINVGGKYVKLQIWDTAGQERFRSVTRSYYR 84
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L NWL DAR LA +I I + GNK D+ R VTFLEASRF
Sbjct 85 GAAGALLVYDITSRETYNALTNWLTDARMLASQNIVIILCGNKKDLDADREVTFLEASRF 144
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + FV+ AR IL+KI+ G
Sbjct 145 AQENELMFLETSALTGENVEEAFVQCARKILNKIESG 181
> mmu:19341 Rab4a, AI848268, Rab4; RAB4A, member RAS oncogene
family; K07879 Ras-related protein Rab-4A
Length=218
Score = 224 bits (570), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 103/157 (65%), Positives = 128/157 (81%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+E KF+ S+HTIGVEF SK++N+ +KLQIWDTAGQER+R+VTRSYYR
Sbjct 25 GKSCLLHQFIEKKFKDDSNHTIGVEFGSKIINVGGKYVKLQIWDTAGQERFRSVTRSYYR 84
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L NWL DAR LA +I + + GNK D+ R VTFLEASRF
Sbjct 85 GAAGALLVYDITSRETYNALTNWLTDARMLASQNIVLILCGNKKDLDADREVTFLEASRF 144
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + F++ AR IL+KI+ G
Sbjct 145 AQENELMFLETSALTGENVEEAFMQCARKILNKIESG 181
> dre:403031 rab4a, fc93b03, wu:fc93b03; RAB4a, member RAS oncogene
family; K07879 Ras-related protein Rab-4A
Length=213
Score = 221 bits (563), Expect = 8e-58, Method: Compositional matrix adjust.
Identities = 103/157 (65%), Positives = 129/157 (82%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF+E +F+ S+HTIGVEF SK++++ + +KLQIWDTAGQER+R+VTRSYYR
Sbjct 20 GKSCLLHQFIEKRFKDDSNHTIGVEFGSKIISVVNKFVKLQIWDTAGQERFRSVTRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ +RE+YN L NWL DAR LA +I I + GNK D+ R VTFLEASRF
Sbjct 80 GAAGALLVYDITSRETYNALTNWLTDARMLASQNIVIILCGNKKDLDADREVTFLEASRF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
AQEN+++FLETSALTGE V + FV+ AR IL+KI+ G
Sbjct 140 AQENELMFLETSALTGENVEEAFVQCARKILNKIESG 176
> dre:393656 rab14l, MGC63643, zgc:63643; RAB14, member RAS oncogene
family, like; K07881 Ras-related protein Rab-14
Length=215
Score = 206 bits (524), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 93/157 (59%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ KIKLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIMEVSGQKIKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLGSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> xla:494665 rab14; RAB14, member RAS oncogene family; K07881
Ras-related protein Rab-14
Length=215
Score = 206 bits (524), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 93/157 (59%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ KIKLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKIKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> mmu:68365 Rab14, 0610030G24Rik, 2810475J17Rik, A830021G03Rik,
AI314285, AI649155, D030017L14Rik; RAB14, member RAS oncogene
family; K07881 Ras-related protein Rab-14
Length=215
Score = 206 bits (524), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 93/157 (59%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ KIKLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKIKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> hsa:51552 RAB14, FBP, RAB-14; RAB14, member RAS oncogene family;
K07881 Ras-related protein Rab-14
Length=215
Score = 206 bits (524), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 93/157 (59%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ KIKLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKIKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> dre:373104 rab14, cb731, fc26g11, fj47f07, fj68a01, wu:fc26g11,
wu:fj47f07, wu:fj68a01; RAB14, member RAS oncogene family;
K07881 Ras-related protein Rab-14
Length=215
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 92/157 (58%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ K+KLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKVKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> dre:100332590 RAB14, member RAS oncogene family-like
Length=215
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 92/157 (58%), Positives = 117/157 (74%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ K+KLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKVKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WL DAR+L + I ++GNK D+ +R VT+ EA +F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLTDARNLTNPNTVIILIGNKADLEAQRDVTYEEAKQF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN +LFLE SA TGE V D F+ A+ I IQDG
Sbjct 143 AEENGLLFLEASAKTGENVEDAFLEAAKKIYQNIQDG 179
> cel:K09A9.2 rab-14; RAB family member (rab-14); K07881 Ras-related
protein Rab-14
Length=210
Score = 205 bits (521), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 92/157 (58%), Positives = 116/157 (73%), Gaps = 0/157 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLLHQF E KF HTIGVEF ++++ ++ KIKLQIWDTAGQER+RAVTRSYYR
Sbjct 23 GKSCLLHQFTEKKFMADCPHTIGVEFGTRIIEVSGQKIKLQIWDTAGQERFRAVTRSYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA++VYD+ R +YNHL +WLADA+SL + +I ++GNK D+ D+R V + EA F
Sbjct 83 GAAGALMVYDITRRSTYNHLSSWLADAKSLTNPNTAIFLIGNKADLEDQRDVPYEEAKAF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDG 157
A+EN + FLE SA TG V D F+ A+ I IQDG
Sbjct 143 AEENGLTFLECSAKTGSNVEDAFLETAKQIYQNIQDG 179
> ath:AT4G17170 RABB1C; RABB1C (ARABIDOPSIS RAB GTPASE HOMOLOG
B1C); GTP binding / GTPase; K07976 Rab family, other
Length=211
Score = 182 bits (463), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 81/158 (51%), Positives = 118/158 (74%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ +++ IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ RE++NHL +WL DAR A A+++I ++GNK D+ +RAV+ E +F
Sbjct 78 GAAGALLVYDITRRETFNHLASWLEDARQHANANMTIMLIGNKCDLAHRRAVSTEEGEQF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+E SA T + V + F++ A I KIQDG+
Sbjct 138 AKEHGLIFMEASAKTAQNVEEAFIKTAATIYKKIQDGV 175
> cpv:cgd1_2060 Rab2 GTPase
Length=185
Score = 174 bits (442), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 84/158 (53%), Positives = 114/158 (72%), Gaps = 1/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +FR TIGVEF ++++NL+ KIKLQIWDTAGQE +R++TRSYYR
Sbjct 20 GKSCLLLQFTDRRFRVDHDLTIGVEFGARIINLDAKKIKLQIWDTAGQESFRSITRSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++NHL WL D + A +++I +VGNK D+ D+R VT E F
Sbjct 80 GAAGALLVYDITRRDTFNHLSKWLNDVKRNATPNMTIILVGNKSDL-DRREVTTEEGVEF 138
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A++N +LF+ETSA T V + F++VA I I DG+
Sbjct 139 AEKNGLLFIETSAKTSNNVEEAFMKVAEKIYKNILDGI 176
> ath:AT4G17160 ATRABB1A (ARABIDOPSIS RAB GTPASE HOMOLOG B1A);
GTP binding; K07976 Rab family, other
Length=205
Score = 173 bits (439), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 78/158 (49%), Positives = 111/158 (70%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL +F + +F+ TIGVEF +K + +++ IKLQIWDTAGQE +R+VTRSYYR
Sbjct 18 GKSCLLLKFTDKRFQAVHDLTIGVEFGAKTITIDNKPIKLQIWDTAGQESFRSVTRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
G AG +LVYD+ RE++NHL +WL +AR A +++ ++GNK D+ DKR V+ E +F
Sbjct 78 GRAGTLLVYDITRRETFNHLASWLEEARQHASENMTTMLIGNKCDLEDKRTVSTEEGEQF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+E SA T V + FV A I +IQDG+
Sbjct 138 AREHGLIFMEASAKTAHNVEEAFVETAATIYKRIQDGV 175
> xla:443746 rab2b, MGC132078, MGC78967; rab2b, member RAS oncogene
family; K07878 Ras-related protein Rab-2B
Length=215
Score = 172 bits (437), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 79/158 (50%), Positives = 114/158 (72%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++N++ IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMINIDGKPIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ RE+++HL +WL DAR + +++ I ++GNK D+ +R V+ E F
Sbjct 78 GAAGALLVYDITRRETFSHLTSWLEDARQHSSSNMVIILIGNKSDLESRRDVSREEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + F+ A+ I KIQ GL
Sbjct 138 AREHGLIFMETSAKTSANVEEAFIDTAKEIYKKIQQGL 175
> mmu:76338 Rab2b, 1500012D09Rik, 4930528G15Rik, A230002G14, D530043M21Rik;
RAB2B, member RAS oncogene family; K07878 Ras-related
protein Rab-2B
Length=216
Score = 172 bits (437), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 79/158 (50%), Positives = 114/158 (72%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++N++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMVNIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ RE++NHL +WL DAR + +++ I ++GNK D+ +R V E F
Sbjct 78 GAAGALLVYDITRRETFNHLTSWLEDARQHSSSNMVIMLIGNKSDLESRRDVKREEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + ++ A+ I KIQ GL
Sbjct 138 AREHGLIFMETSAKTACNVEEAYINTAKEIYRKIQQGL 175
> dre:140617 rab2a, cb75, fb57e06, rab2, wu:fb57e06; RAB2A, member
RAS oncogene family; K07877 Ras-related protein Rab-2A
Length=212
Score = 172 bits (435), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 77/158 (48%), Positives = 113/158 (71%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ ++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++NHL WL DAR + +++ I ++GNK D+ +R V E F
Sbjct 78 GAAGALLVYDITRRDTFNHLTTWLEDARQHSNSNMVIMLIGNKSDLESRREVKKEEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + F+ A+ I KIQ+G+
Sbjct 138 AREHGLIFMETSAKTASNVEEAFINTAKEIYEKIQEGV 175
> mmu:59021 Rab2a, 9330148M11Rik, C80220, Rab2; RAB2A, member
RAS oncogene family; K07877 Ras-related protein Rab-2A
Length=212
Score = 171 bits (434), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 77/158 (48%), Positives = 113/158 (71%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ ++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++NHL WL DAR + +++ I ++GNK D+ +R V E F
Sbjct 78 GAAGALLVYDITRRDTFNHLTTWLEDARQHSNSNMVIMLIGNKSDLESRREVKKEEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + F+ A+ I KIQ+G+
Sbjct 138 AREHGLIFMETSAKTASNVEEAFINTAKEIYEKIQEGV 175
> hsa:5862 RAB2A, RAB2; RAB2A, member RAS oncogene family; K07877
Ras-related protein Rab-2A
Length=212
Score = 171 bits (434), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 77/158 (48%), Positives = 113/158 (71%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ ++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++NHL WL DAR + +++ I ++GNK D+ +R V E F
Sbjct 78 GAAGALLVYDITRRDTFNHLTTWLEDARQHSNSNMVIMLIGNKSDLESRREVKKEEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + F+ A+ I KIQ+G+
Sbjct 138 AREHGLIFMETSAKTASNVEEAFINTAKEIYEKIQEGV 175
> cel:F53F10.4 unc-108; UNCoordinated family member (unc-108);
K07878 Ras-related protein Rab-2B
Length=214
Score = 169 bits (428), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 77/158 (48%), Positives = 114/158 (72%), Gaps = 0/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ ++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMVTIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++NHL +WL DAR + +++ I ++GNK D+ +R V E F
Sbjct 78 GAAGALLVYDITRRDTFNHLTSWLEDARQHSNSNMVIMLIGNKSDLEARREVKREEGEAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+E+ ++F+ETSA T V + F+ A+ I KIQ+G+
Sbjct 138 AREHGLVFMETSAKTAANVEEAFIDTAKEIYRKIQEGV 175
> pfa:PFL1500w rab2; Rab2, GTPase (EC:3.6.1.-); K07976 Rab family,
other
Length=213
Score = 167 bits (424), Expect = 9e-42, Method: Compositional matrix adjust.
Identities = 79/154 (51%), Positives = 113/154 (73%), Gaps = 1/154 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +FR TIGVEF ++++NL++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 19 GKSCLLLQFTDKRFRADHDLTIGVEFGARLINLDNKQIKLQIWDTAGQESFRSITRSYYR 78
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ RE++NHL WL + R + ++I +VGNK D+ ++R V+ E ++F
Sbjct 79 GAAGALLVYDITRRETFNHLNRWLDEVRQNSNPHMAIILVGNKCDL-ERREVSAEEGAQF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKI 154
A++N ++FLETSA T + V + F+ AR I I
Sbjct 138 ARQNGLIFLETSAKTAKNVEEAFLYTARKIYDNI 171
> tgo:TGME49_112050 Ras family domain-containing protein (EC:3.1.4.12)
Length=214
Score = 162 bits (409), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 75/158 (47%), Positives = 113/158 (71%), Gaps = 1/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +FR TIGVEF ++++++ ++KLQIWDTAGQE +R++TRSYYR
Sbjct 19 GKSCLLLQFTDKRFRTDHDLTIGVEFGARLVSIAGRQVKLQIWDTAGQESFRSITRSYYR 78
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LVYD+ R+++ HL WL + R + ++I ++GNK D+ ++R V+F E + F
Sbjct 79 GAAGALLVYDITRRDTFLHLTRWLDEVRQNSNPHMTIMLIGNKSDL-ERREVSFDEGAAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A+++ ++FLETSA T + V + F+ AR I IQ G+
Sbjct 138 ARQHGLIFLETSAKTAQNVDEAFILTARKIYENIQRGI 175
> xla:735215 rab2a, MGC131016, rab2; RAB2A, member RAS oncogene
family; K07877 Ras-related protein Rab-2A
Length=228
Score = 158 bits (399), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 76/175 (43%), Positives = 112/175 (64%), Gaps = 17/175 (9%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL QF + +F+ TIGVEF ++++ ++ +IKLQIWDTAGQE +R++TRSYYR
Sbjct 18 GKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDGKQIKLQIWDTAGQESFRSITRSYYR 77
Query 61 -----------------GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNK 103
GAAGA+LVYD+ R+++NHL WL DAR + +++ I ++ NK
Sbjct 78 GAAGALLVYDITRSYYRGAAGALLVYDITRRDTFNHLTTWLEDARQHSNSNMVIMLIANK 137
Query 104 IDVRDKRAVTFLEASRFAQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
D+ +R V E FA+E+ ++F+ETSA T V + F+ A+ I KIQ+G+
Sbjct 138 SDLESRREVKKEEGEAFAREHGLVFMETSAKTASNVEEAFINTAKEIYEKIQEGV 192
> sce:YER031C YPT31, YPT8; GTPase of the Ypt/Rab family, very
similar to Ypt32p; involved in the exocytic pathway; mediates
intra-Golgi traffic or the budding of post-Golgi vesicles
from the trans-Golgi; K07976 Rab family, other
Length=223
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 74/154 (48%), Positives = 103/154 (66%), Gaps = 0/154 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F +N+F S TIGVEFA++ L ++ +IK QIWDTAGQERYRA+T +YYR
Sbjct 25 GKSNLLSRFTKNEFNMDSKSTIGVEFATRTLEIDGKRIKAQIWDTAGQERYRAITSAYYR 84
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA++VYD++ SY + +WL++ R A ++++ ++GNK D+ RAV E+ F
Sbjct 85 GAVGALIVYDISKSSSYENCNHWLSELRENADDNVAVGLIGNKSDLAHLRAVPTEESKTF 144
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKI 154
AQEN +LF ETSAL E V F + I K+
Sbjct 145 AQENQLLFTETSALNSENVDKAFEELINTIYQKV 178
> tpv:TP01_0877 hypothetical protein; K07976 Rab family, other
Length=215
Score = 155 bits (392), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 72/158 (45%), Positives = 105/158 (66%), Gaps = 1/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL +F + +F+ TIGVEF ++ + + +IKLQIWDTAGQE +R++TRSYYR
Sbjct 19 GKSCLLLRFTDKRFKPDHDLTIGVEFGTRFITVQGEQIKLQIWDTAGQESFRSITRSYYR 78
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LV+D++ R ++ HL WL + R +SI ++GNK DV R V + E +F
Sbjct 79 GAAGALLVFDISRRSTFEHLTKWLEEVRDHGSPTMSIILIGNKTDVA-SREVEYHEGKQF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A +ND++++ETSA V F+ A LI +++GL
Sbjct 138 ADQNDLIYMETSAKGDYNVEAAFLTTAELIYENVKNGL 175
> bbo:BBOV_IV008990 23.m06418; Rab2; K07976 Rab family, other
Length=217
Score = 154 bits (388), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 70/158 (44%), Positives = 106/158 (67%), Gaps = 1/158 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL +F + +F+ TIGVEF ++++N+ IKLQIWDTAGQE +R++TRSYYR
Sbjct 19 GKSCLLLRFTDKRFKPDHDLTIGVEFGTRLINVKGDTIKLQIWDTAGQESFRSITRSYYR 78
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GAAGA+LV+D++ R ++ HL WL + + ++I ++GNK DV +KR V + E F
Sbjct 79 GAAGALLVFDISRRSTFEHLTRWLDEVKDHGSPTMTIVLIGNKTDV-NKREVEYHEGKAF 137
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQDGL 158
A EN+++++ETSA V + F+ A +I + G+
Sbjct 138 ADENNLIYMETSAKGDYNVEEAFLTTATMIYDNVNRGM 175
> ath:AT4G35860 ATGB2; ATGB2 (GTP-BINDING 2); GTP binding; K07976
Rab family, other
Length=165
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 69/129 (53%), Positives = 95/129 (73%), Gaps = 0/129 (0%)
Query 30 VLNLNDMKIKLQIWDTAGQERYRAVTRSYYRGAAGAILVYDVANRESYNHLINWLADARS 89
++ ++ IKLQIWDTAGQE +R++TRSYYRGAAGA+LVYD+ RE++NHL +WL DAR
Sbjct 1 MVTVDGRPIKLQIWDTAGQESFRSITRSYYRGAAGALLVYDITRRETFNHLASWLEDARQ 60
Query 90 LARADISIAIVGNKIDVRDKRAVTFLEASRFAQENDVLFLETSALTGEGVTDVFVRVARL 149
A ++SI ++GNK D+ KRAV+ E +FA+E+ +LFLE SA T + V + F+ A
Sbjct 61 HANPNMSIMLIGNKCDLAHKRAVSKEEGQQFAKEHGLLFLEASARTAQNVEEAFIETAAK 120
Query 150 ILSKIQDGL 158
IL IQDG+
Sbjct 121 ILQNIQDGV 129
> bbo:BBOV_IV006550 23.m06035; ras-related protein Rab11A; K07976
Rab family, other
Length=212
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 80/157 (50%), Positives = 105/157 (66%), Gaps = 5/157 (3%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKI-KLQIWDTAGQERYRAVTRSYY 59
GKS LL +FV+ F+ S TIGVEFA+K + LN+ KI K QIWDTAGQERYRA+T +YY
Sbjct 22 GKSNLLDRFVKGNFKLDSKSTIGVEFATKTVTLNNGKIAKAQIWDTAGQERYRAITSAYY 81
Query 60 RGAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASR 119
RGA GAI+VYD+A + S+ ++ WL + A ++I+I +VGNK D+ R V +
Sbjct 82 RGAMGAIIVYDIACKTSFTNVSKWLTELHDYADSNITICLVGNKSDLTHLREVNKEDGEA 141
Query 120 FAQENDVLFLETSALTGEGVTDVFVRVARLILSKIQD 156
FA+END+LF ETS L E V F + +LSKI D
Sbjct 142 FARENDLLFFETSCLNSENVDVAF----KELLSKIGD 174
> ath:AT4G39990 RABA4B; RABA4B (RAB GTPASE HOMOLOG A4B); GTP binding;
K07976 Rab family, other
Length=224
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 72/143 (50%), Positives = 97/143 (67%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F ++F S TIGVEF ++ L++ IK QIWDTAGQERYRAVT +YYR
Sbjct 29 GKSQLLARFARDEFSMDSKATIGVEFQTRTLSIEQKSIKAQIWDTAGQERYRAVTSAYYR 88
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+ RE++ H+ WL + R+ A +I I ++GNK D+ D+RAV +A F
Sbjct 89 GAVGAMLVYDMTKRETFEHIPRWLEELRAHADKNIVIILIGNKSDLEDQRAVPTEDAKEF 148
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++ + FLETSAL V + F
Sbjct 149 AEKEGLFFLETSALNATNVENSF 171
> tpv:TP01_1204 hypothetical protein; K07976 Rab family, other
Length=212
Score = 149 bits (377), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 73/151 (48%), Positives = 106/151 (70%), Gaps = 1/151 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKI-KLQIWDTAGQERYRAVTRSYY 59
GKS LL +FV+ F+ S TIGVEFA+K +NL + K+ K QIWDTAGQERYRA+T +YY
Sbjct 22 GKSNLLDRFVKGNFKLDSKSTIGVEFATKNVNLRNGKVAKAQIWDTAGQERYRAITSAYY 81
Query 60 RGAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASR 119
RGA GAI+VYD+A+++S+ ++ WL++ A++ IA+VGNK D+ R VT+ + R
Sbjct 82 RGARGAIVVYDIASKQSFYNVSRWLSELNEYGDANMIIALVGNKSDLTHLREVTYEDGER 141
Query 120 FAQENDVLFLETSALTGEGVTDVFVRVARLI 150
+A+ N+++F ETS L+ E + F + LI
Sbjct 142 YAKSNNLIFFETSCLSNENIDTTFTELLNLI 172
> ath:AT3G12160 RABA4D; RABA4D (RAB GTPASE HOMOLOG A4D); GTP binding;
K07976 Rab family, other
Length=222
Score = 148 bits (373), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 71/146 (48%), Positives = 97/146 (66%), Gaps = 0/146 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GK+ LL +F N+F S TIGVEF +K L +++ +K QIWDTAGQERYRAVT +YYR
Sbjct 27 GKTQLLARFARNEFSVDSKATIGVEFQTKTLVIDNKTVKAQIWDTAGQERYRAVTSAYYR 86
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+ R+S++H+ WL + R A +I I ++GNK D+ RAV +A F
Sbjct 87 GAVGAMLVYDMTKRQSFDHMAKWLEELRGHADKNIVIMLIGNKCDLGSLRAVPTEDAQEF 146
Query 121 AQENDVLFLETSALTGEGVTDVFVRV 146
AQ ++ F+ETSAL V F+ +
Sbjct 147 AQRENLFFMETSALEATNVETAFLTI 172
> ath:AT5G47960 ATRABA4C; GTP binding; K07976 Rab family, other
Length=223
Score = 147 bits (371), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 73/146 (50%), Positives = 96/146 (65%), Gaps = 0/146 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEF ++ L ++ IK QIWDTAGQERYRAVT +YYR
Sbjct 27 GKSQLLARFSRNEFSIESKATIGVEFQTRTLEIDRKTIKAQIWDTAGQERYRAVTSAYYR 86
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+ R+S++H+ WL + R A +I I ++GNK D+ RAV +A F
Sbjct 87 GAVGAMLVYDITKRQSFDHVARWLEELRGHADKNIVIMLIGNKTDLGTLRAVPTEDAKEF 146
Query 121 AQENDVLFLETSALTGEGVTDVFVRV 146
AQ ++ F+ETSAL V F+ V
Sbjct 147 AQRENLFFMETSALDSNNVEPSFLTV 172
> sce:YGL210W YPT32; GTPase of the Ypt/Rab family, very similar
to Ypt31p; involved in the exocytic pathway; mediates intra-Golgi
traffic or the budding of post-Golgi vesicles from the
trans-Golgi; K07976 Rab family, other
Length=222
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 70/143 (48%), Positives = 96/143 (67%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F ++F S TIGVEFA++ + + + KIK QIWDTAGQERYRA+T +YYR
Sbjct 25 GKSNLLSRFTTDEFNIESKSTIGVEFATRTIEVENKKIKAQIWDTAGQERYRAITSAYYR 84
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA++VYD++ SY + +WL + R A ++++ ++GNK D+ RAV EA F
Sbjct 85 GAVGALIVYDISKSSSYENCNHWLTELRENADDNVAVGLIGNKSDLAHLRAVPTDEAKNF 144
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A EN +LF ETSAL + V F
Sbjct 145 AMENQMLFTETSALNSDNVDKAF 167
> hsa:9230 RAB11B, H-YPT3, MGC133246; RAB11B, member RAS oncogene
family; K07905 Ras-related protein Rab-11B
Length=218
Score = 145 bits (366), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 98/143 (68%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ IK QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTIKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A ++I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADSNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++N++ F+ETSAL V + F
Sbjct 143 AEKNNLSFIETSALDSTNVEEAF 165
> mmu:19326 Rab11b, A730055L17Rik; RAB11B, member RAS oncogene
family; K07905 Ras-related protein Rab-11B
Length=218
Score = 145 bits (366), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 98/143 (68%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ IK QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTIKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A ++I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADSNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++N++ F+ETSAL V + F
Sbjct 143 AEKNNLSFIETSALDSTNVEEAF 165
> tgo:TGME49_014770 RAS small GTpase, putative (EC:3.1.4.12);
K07874 Ras-related protein Rab-1A
Length=202
Score = 145 bits (365), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 70/155 (45%), Positives = 100/155 (64%), Gaps = 0/155 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL +F ++ + S TIGV+F + ++L+ +KLQIWDTAGQER+R +T SYYR
Sbjct 20 GKSCLLLRFADDTYTESYISTIGVDFKIRTIDLDGKTVKLQIWDTAGQERFRTITSSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA G I+VYDV +RES+N++ NW+ + A +S +VGNK D+ KR VT+ E F
Sbjct 80 GAHGIIIVYDVTDRESFNNVKNWMMEIDKYAMEGVSKLLVGNKCDLTSKRTVTYEEGKEF 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQ 155
A ++ F+ETSA V F +A I +++Q
Sbjct 140 ADSCNMRFIETSAKNAHNVEQAFHIMASEIKARVQ 174
> pfa:PFE0625w Rab1b; Rab1b, GTPase; K07976 Rab family, other
Length=200
Score = 144 bits (364), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 70/155 (45%), Positives = 99/155 (63%), Gaps = 0/155 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSCLL +F ++ + S TIGV+F K + + D IKLQIWDTAGQER+R +T SYYR
Sbjct 20 GKSCLLLRFADDTYTDSYISTIGVDFKIKTIEIEDKIIKLQIWDTAGQERFRTITSSYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA G I+VYDV +R+S+N++ NW+ + A D+ ++GNKID+++ R V++ E
Sbjct 80 GAQGIIIVYDVTDRDSFNNVKNWIIEIEKYASEDVQKILIGNKIDLKNDRNVSYEEGKEL 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQ 155
A ++ FLETSA V F +A I +K Q
Sbjct 140 ADSCNIQFLETSAKIAHNVEQAFKTMAYEIKNKSQ 174
> pfa:PFA_0335w Rab5c; Rab5c, GTPase (EC:3.6.5.2); K07976 Rab
family, other
Length=214
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 64/148 (43%), Positives = 102/148 (68%), Gaps = 0/148 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKSC++ +F +N+F TIG F ++++++ + IK +IWDTAGQERYR++ YYR
Sbjct 39 GKSCIVVRFAKNEFYEYQESTIGAAFMTQLIDIGECTIKFEIWDTAGQERYRSLAPMYYR 98
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA+ A++VYD+ N++S+ W+ + +S+ DI IA+ GNK D+ + RAV A F
Sbjct 99 GASAAVIVYDITNKKSFEGAKGWIHELKSVHSNDIIIALAGNKNDLEEHRAVDRELAESF 158
Query 121 AQENDVLFLETSALTGEGVTDVFVRVAR 148
A N++LF+ETSA TG+ V ++F+R+A+
Sbjct 159 ANSNNILFIETSAKTGQNVNELFLRIAK 186
> dre:492487 rab11a, wu:fi15h09, zgc:103679; RAB11a, member RAS
oncogene family; K07904 Ras-related protein Rab-11A
Length=215
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 76/154 (49%), Positives = 102/154 (66%), Gaps = 4/154 (2%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ +K QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTVKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A ++I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADSNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKI 154
A++N + FLETSAL V F + IL++I
Sbjct 143 AEKNGLSFLETSALDSTNVETAF----QTILTEI 172
> xla:494642 hypothetical LOC494642; K07905 Ras-related protein
Rab-11B
Length=218
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 97/143 (67%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ IK QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTIKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A +I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADNNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++N++ F+ETSAL V + F
Sbjct 143 AEKNNLSFIETSALDSTNVEEAF 165
> dre:436828 zgc:92772; K07905 Ras-related protein Rab-11B
Length=218
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 97/143 (67%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ IK QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTIKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A +I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADNNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++N++ F+ETSAL V + F
Sbjct 143 AEKNNLSFIETSALDSTNVEEAF 165
> dre:100093706 rab8b, zgc:165529; RAB8B, member RAS oncogene
family; K07902 Ras-related protein Rab-8B
Length=209
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 67/155 (43%), Positives = 103/155 (66%), Gaps = 0/155 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GK+CLL +F E+ F + TIG++F + + L+ KIKLQIWDTAGQER+R +T +YYR
Sbjct 20 GKTCLLFRFSEDAFNTTFISTIGIDFKIRTIELDGKKIKLQIWDTAGQERFRTITTAYYR 79
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA G +LVYD+ N +S++++ NW+ + A +D+ I+GNK D+ DKR V+ +
Sbjct 80 GAMGIMLVYDITNEKSFDNIKNWIRNIEEHASSDVERMILGNKCDMNDKRQVSKERGEKL 139
Query 121 AQENDVLFLETSALTGEGVTDVFVRVARLILSKIQ 155
A + + FLETSA + V + FV +AR I++++
Sbjct 140 AIDYGIKFLETSAKSSTNVEEAFVTLARDIMTRLN 174
> dre:406798 rab11b, wu:fb07g11, zgc:55760; RAB11B, member RAS
oncogene family; K07905 Ras-related protein Rab-11B
Length=218
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 73/143 (51%), Positives = 97/143 (67%), Gaps = 0/143 (0%)
Query 1 GKSCLLHQFVENKFRRSSSHTIGVEFASKVLNLNDMKIKLQIWDTAGQERYRAVTRSYYR 60
GKS LL +F N+F S TIGVEFA++ + ++ IK QIWDTAGQERYRA+T +YYR
Sbjct 23 GKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTIKAQIWDTAGQERYRAITSAYYR 82
Query 61 GAAGAILVYDVANRESYNHLINWLADARSLARADISIAIVGNKIDVRDKRAVTFLEASRF 120
GA GA+LVYD+A +Y ++ WL + R A +I I +VGNK D+R RAV EA F
Sbjct 83 GAVGALLVYDIAKHLTYENVERWLKELRDHADNNIVIMLVGNKSDLRHLRAVPTDEARAF 142
Query 121 AQENDVLFLETSALTGEGVTDVF 143
A++N++ F+ETSAL V + F
Sbjct 143 AEKNNLSFIETSALDSTNVEEAF 165
> hsa:84932 RAB2B, FLJ14824; RAB2B, member RAS oncogene family;
K07878 Ras-related protein Rab-2B
Length=170
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 65/129 (50%), Positives = 94/129 (72%), Gaps = 0/129 (0%)
Query 30 VLNLNDMKIKLQIWDTAGQERYRAVTRSYYRGAAGAILVYDVANRESYNHLINWLADARS 89
++N++ +IKLQIWDTAGQE +R++TRSYYRGAAGA+LVYD+ RE++NHL +WL DAR
Sbjct 1 MVNIDGKQIKLQIWDTAGQESFRSITRSYYRGAAGALLVYDITRRETFNHLTSWLEDARQ 60
Query 90 LARADISIAIVGNKIDVRDKRAVTFLEASRFAQENDVLFLETSALTGEGVTDVFVRVARL 149
+ +++ I ++GNK D+ +R V E FA+E+ ++F+ETSA T V + F+ A+
Sbjct 61 HSSSNMVIMLIGNKSDLESRRDVKREEGEAFAREHGLIFMETSAKTACNVEEAFINTAKE 120
Query 150 ILSKIQDGL 158
I KIQ GL
Sbjct 121 IYRKIQQGL 129
Lambda K H
0.322 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40