bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1693_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
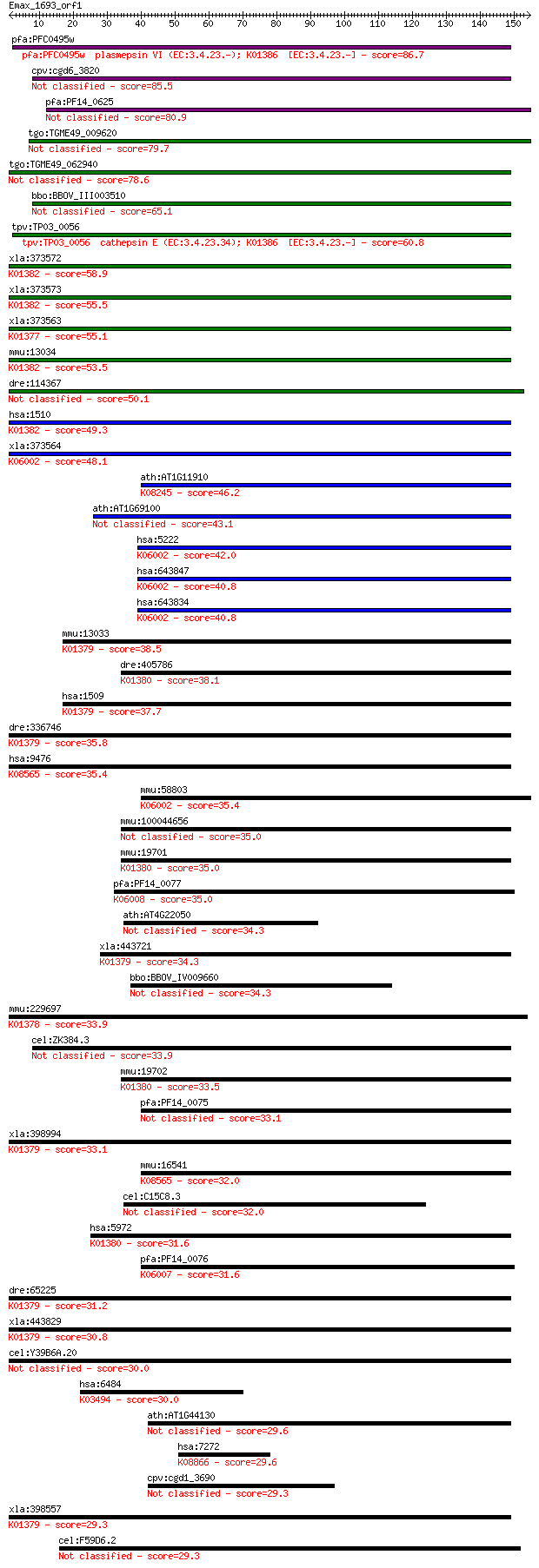
pfa:PFC0495w plasmepsin VI (EC:3.4.23.-); K01386 [EC:3.4.23.-] 86.7 2e-17
cpv:cgd6_3820 membrane bound aspartyl proteinase with a signal... 85.5 6e-17
pfa:PF14_0625 plasmepsin VIII (EC:3.4.17.4) 80.9 2e-15
tgo:TGME49_009620 eukaryotic aspartyl protease, putative (EC:3... 79.7 4e-15
tgo:TGME49_062940 eukaryotic aspartyl protease, putative (EC:3... 78.6 7e-15
bbo:BBOV_III003510 17.m07331; eukaryotic aspartyl protease fam... 65.1 8e-11
tpv:TP03_0056 cathepsin E (EC:3.4.23.34); K01386 [EC:3.4.23.-] 60.8 2e-09
xla:373572 ctse-a, CE1, cate, ce1-A, ce2, ctse; cathepsin E (E... 58.9 7e-09
xla:373573 ctse-b, CE2, cate, ce1, ce2-A; cathepsin E (EC:3.4.... 55.5 8e-08
xla:373563 pgc-A; pepsinogen C (EC:3.4.23.3); K01377 gastricsi... 55.1 9e-08
mmu:13034 Ctse, A430072O03Rik, C920004C08Rik, CE, CatE; cathep... 53.5 3e-07
dre:114367 nots, LAP, MGC192423, ctsel, zgc:103608; nothepsin 50.1 3e-06
hsa:1510 CTSE, CATE; cathepsin E (EC:3.4.23.34); K01382 cathep... 49.3 5e-06
xla:373564 pga4, pga-A; pepsinogen 4, group I (pepsinogen A); ... 48.1 1e-05
ath:AT1G11910 aspartyl protease family protein; K08245 phyteps... 46.2 4e-05
ath:AT1G69100 aspartic-type endopeptidase 43.1 3e-04
hsa:5222 PGA5, PGA3, PGA4; pepsinogen 5, group I (pepsinogen A... 42.0 7e-04
hsa:643847 PGA4, FLJ58952, FLJ77962, PGA3, PGA5; pepsinogen 4,... 40.8 0.002
hsa:643834 PGA3, DKFZp666J2410, PGA4, PGA5; pepsinogen 3, grou... 40.8 0.002
mmu:13033 Ctsd, CD, CatD; cathepsin D (EC:3.4.23.5); K01379 ca... 38.5 0.009
dre:405786 ren; renin (EC:3.4.23.15); K01380 renin [EC:3.4.23.15] 38.1 0.010
hsa:1509 CTSD, CLN10, CPSD, MGC2311; cathepsin D (EC:3.4.23.5)... 37.7 0.014
dre:336746 fa94d12, wu:fa94d12; zgc:63831; K01379 cathepsin D ... 35.8 0.055
hsa:9476 NAPSA, KAP, Kdap, NAP1, NAPA, SNAPA; napsin A asparti... 35.4 0.081
mmu:58803 Pga5, 1110035E17Rik, Pepf; pepsinogen 5, group I (EC... 35.4 0.082
mmu:100044656 renin-1-like 35.0 0.089
mmu:19701 Ren1, D19352, Ren, Ren-1, Ren-A, Ren1c, Ren1d, Rn-1,... 35.0 0.10
pfa:PF14_0077 plasmepsin II; K06008 plasmepsin II [EC:3.4.23.39] 35.0 0.10
ath:AT4G22050 aspartyl protease family protein 34.3 0.15
xla:443721 ctsd; cathepsin D (EC:3.4.23.5); K01379 cathepsin D... 34.3 0.16
bbo:BBOV_IV009660 23.m06138; aspartyl protease 34.3 0.17
mmu:229697 Cym, Gm131; chymosin (EC:3.4.23.4); K01378 chymosin... 33.9 0.22
cel:ZK384.3 hypothetical protein 33.9 0.24
mmu:19702 Ren2, Ren, Ren-2, Ren-B, Rn-2, Rnr; renin 2 tandem d... 33.5 0.31
pfa:PF14_0075 plasmepsin IV 33.1 0.32
xla:398994 napsa, MGC68767, kap, kdap, nap1, napa, snapa; naps... 33.1 0.37
mmu:16541 Napsa, KAP, Kdap, NAP1, SNAPA, pronapsin; napsin A a... 32.0 0.76
cel:C15C8.3 hypothetical protein 32.0 0.84
hsa:5972 REN, FLJ10761, HNFJ2; renin (EC:3.4.23.15); K01380 re... 31.6 0.96
pfa:PF14_0076 plasmepsin I; K06007 plasmepsin I [EC:3.4.23.38] 31.6 1.1
dre:65225 ctsd, catD, fb93e11, fj17b09, wu:fb93e11, wu:fj17b09... 31.2 1.5
xla:443829 MGC82347 protein; K01379 cathepsin D [EC:3.4.23.5] 30.8 1.9
cel:Y39B6A.20 asp-1; ASpartyl Protease family member (asp-1) 30.0 3.3
hsa:6484 ST3GAL4, CGS23, FLJ11867, FLJ46764, NANTA3, SAT3, SIA... 30.0 3.3
ath:AT1G44130 nucellin protein, putative 29.6 3.5
hsa:7272 TTK, CT96, ESK, FLJ38280, MPS1, MPS1L1, PYT; TTK prot... 29.6 4.1
cpv:cgd1_3690 aspartyl (acid) protease 29.3 4.6
xla:398557 cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3... 29.3 4.7
cel:F59D6.2 hypothetical protein 29.3 4.7
> pfa:PFC0495w plasmepsin VI (EC:3.4.23.-); K01386 [EC:3.4.23.-]
Length=432
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 51/159 (32%), Positives = 83/159 (52%), Gaps = 15/159 (9%)
Query 2 YIDSMTGREVL-------HLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHP----- 49
YI TG +L +L+ +K+ IG + ++ +PFS+LPFDGI GLG
Sbjct 164 YIQYGTGTSILEQSYDDVYLKGLKIKHQCIGLAIEESLHPFSDLPFDGIVGLGFSDPDFR 223
Query 50 HNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLL 109
+ +S LI+ + + +K +F+ +P +G +TFG N+ Y +G +I WFP++
Sbjct 224 SQNKYASPLIETIKKQNLLKRNIFSFYVPKKLEKSGAITFGKANKKYTVEGKSIEWFPVI 283
Query 110 SKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
S WE+ L D+ + + C + C A IDTG+S
Sbjct 284 SLY--YWEINLLDIQLSHKNLFLCESK-KCRAAIDTGSS 319
> cpv:cgd6_3820 membrane bound aspartyl proteinase with a signal
peptide plus transmembrane domain
Length=467
Score = 85.5 bits (210), Expect = 6e-17, Method: Composition-based stats.
Identities = 42/144 (29%), Positives = 79/144 (54%), Gaps = 7/144 (4%)
Query 8 GREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESS---SLIKQLVE 64
G+EV+++ K+K+ D + G + ++ PF+ELPFDG+ GLG P + + ++++ + E
Sbjct 170 GKEVINIGKLKIEDQSFGMAIEESTSPFAELPFDGLVGLGFPDKNSKKNNIPTIVENIKE 229
Query 65 AGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVL 124
+ +F + + +S+ G ++FG + Y G I W L S+ WE+ + D+
Sbjct 230 RNILPRNLFGVYISRDSSTPGSISFGAADPKYTISGQKITWHRLTG--SHYWEIKIKDIK 287
Query 125 VDDATVGFCTPQLPCIAVIDTGTS 148
++ + +C C A IDTG+S
Sbjct 288 INGVSTNYCFGD--CKAAIDTGSS 309
> pfa:PF14_0625 plasmepsin VIII (EC:3.4.17.4)
Length=385
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 77/145 (53%), Gaps = 5/145 (3%)
Query 12 LHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESSSLIKQLVEAGAIKLA 71
+HL +K+ + G ++ PFS++ FDG+ GLG + ++ LI + ++
Sbjct 139 IHLGDIKVKNQEFGLASYISDDPFSDMQFDGLFGLGISDD-KKKKQLIYDSIPKNILEKN 197
Query 72 MFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG 131
MFAI P + G +TFGG+++ +I++ +I WF + S S W + + + ++D +
Sbjct 198 MFAIYYPKNVDDDGAITFGGYDKKFIRENSSIEWFDVTS--SKYWAIQMKGLKINDVFLD 255
Query 132 FCTPQLP--CIAVIDTGTSGDGGDR 154
C+ C AVIDTGTS G +
Sbjct 256 VCSKNHEGFCQAVIDTGTSSIAGPK 280
> tgo:TGME49_009620 eukaryotic aspartyl protease, putative (EC:3.4.23.34)
Length=432
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 58/175 (33%), Positives = 83/175 (47%), Gaps = 29/175 (16%)
Query 7 TGREVLH--LQKVKLHDYAI-----GFVVRQAEYPFSELPFDGIAGLGHPHNVQESSSLI 59
TGR V L+ +++ D AI G VV Q + PF +LPFDGI GLG V L
Sbjct 137 TGRVVYRKALEAIRIGDAAIPSQAVGLVVDQTDEPFVDLPFDGIVGLGEYFVVYTGMCLS 196
Query 60 KQLVEAGA------------------IKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGV 101
+ AG+ I +FA+ L G ++FGGF+ ++Q+G
Sbjct 197 RSSCLAGSYERSGQKDLLDNLKTERVIADKIFAVYLSRRRVMGGVISFGGFDPRFVQQGK 256
Query 102 AIAWFPLLSKQSNRWELWLWDVLVDDATVGFC--TPQLPCIAVIDTGTSGDGGDR 154
I WF L ++ W + L D VD + C + + C+AV+DTGTS GG +
Sbjct 257 NIQWFATLPQEG--WAIPLIDFKVDGVRLHLCFDSAESRCVAVLDTGTSSIGGPK 309
> tgo:TGME49_062940 eukaryotic aspartyl protease, putative (EC:3.4.23.34)
Length=469
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 45/161 (27%), Positives = 84/161 (52%), Gaps = 16/161 (9%)
Query 1 GYIDSMTGREVLHLQK-------VKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQ 53
+I TG VL + K +++ + +G ++++ +PF++LPFDG+ GLG P
Sbjct 167 AFIQYGTGACVLRMAKDTVSIGGIRVQNQTLGLALQESVHPFADLPFDGLVGLGFPDVAG 226
Query 54 ESS------SLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFP 107
E L+ +++ +K FA+ + + GE+TFG N + +G WFP
Sbjct 227 EEGLPPDALPLVDSMMKQKLLKRNAFAVYMSEDLNQPGEITFGSVNPRHTFEGHKPQWFP 286
Query 108 LLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
++S + WE+ + + ++ ++GFC + C A +DTG+S
Sbjct 287 VISL--DYWEVGVHGLRLNRKSLGFCE-RTRCKAAVDTGSS 324
> bbo:BBOV_III003510 17.m07331; eukaryotic aspartyl protease family
protein
Length=521
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 34/145 (23%), Positives = 76/145 (52%), Gaps = 9/145 (6%)
Query 8 GREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESSS----LIKQLV 63
G + + + +++ + +IG ++++PF +LPFDG+ GLG P + + + L+ ++
Sbjct 223 GSDTVKIGPLEVKNQSIGLATYESDHPFGDLPFDGLVGLGFPDKMYKDTDGMLPLLDNIM 282
Query 64 EAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDV 123
+ + + A + + G ++FG + Y+ G + WFP+++ ++ WE+ + +
Sbjct 283 QQKLLNRNLVAFYMSKDKTQPGSMSFGSIDPRYVLPGHSPWWFPVVA--TDFWEIAMEAI 340
Query 124 LVDDATVGFCTPQLPCIAVIDTGTS 148
L+D + + A IDTG+S
Sbjct 341 LIDGKPMKL---ENKYNAAIDTGSS 362
> tpv:TP03_0056 cathepsin E (EC:3.4.23.34); K01386 [EC:3.4.23.-]
Length=513
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 46/160 (28%), Positives = 75/160 (46%), Gaps = 18/160 (11%)
Query 2 YIDSMTGREVLHLQ-------KVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQE 54
YI TG VL L + + +IG V ++E+PF +LPFDG+ GLG P +
Sbjct 227 YIRYGTGECVLALGFDNVKIGSLNVKHQSIGLSVLESEHPFGDLPFDGLVGLGFPDTELK 286
Query 55 SSSLIKQLVEA--GAIKLAMFAIALPLS----ASSAGEVTFGGFNRHYIQKGVAIAWFPL 108
S + + ++ + L + + S G ++FG + Y+ G WFP+
Sbjct 287 ESKKLTPIFDSIRNQVPLTYSTVNRIFNCYNLTKSPGSLSFGSIDPKYVLPGHKPWWFPV 346
Query 109 LSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ +++ WE+ + +LVD V F A IDTG+S
Sbjct 347 V--KTDYWEIEVSSLLVDGEPVQF---DRKYNAAIDTGSS 381
> xla:373572 ctse-a, CE1, cate, ce1-A, ce2, ctse; cathepsin E
(EC:3.4.23.34); K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 58.9 bits (141), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 40/151 (26%), Positives = 78/151 (51%), Gaps = 10/151 (6%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + + G + + ++ + + + G V + F + FDGI GLG+P V + + +
Sbjct 138 GSLSGVIGIDAVTVEGILVQNQQFGESVSEPGSTFVDAEFDGILGLGYPSIAVGDCTPVF 197
Query 60 KQLVEAGAIKLAMFAIALPLSASSA--GEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ ++L MF++ + + +SA GE+ FGGF+ + W P+ ++ W+
Sbjct 198 DNMIAQNLVELPMFSVYMSRNPNSAVGGELVFGGFDASRFSG--QLNWVPVTNQ--GYWQ 253
Query 118 LWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ L +V + + V FC+ C A++DTGTS
Sbjct 254 IQLDNVQI-NGEVLFCSGG--CQAIVDTGTS 281
> xla:373573 ctse-b, CE2, cate, ce1, ce2-A; cathepsin E (EC:3.4.23.34);
K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 38/151 (25%), Positives = 75/151 (49%), Gaps = 10/151 (6%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + + G + + ++ + + + G V + F + FDGI GLG+P V + +
Sbjct 138 GSLSGVIGIDSVTVEGILVQNQQFGESVSEPGSTFVDASFDGILGLGYPSIAVGGCTPVF 197
Query 60 KQLVEAGAIKLAMFAIALPLSASS--AGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ ++L MF++ + +S GE+ FGGF+ + W P+ ++ W+
Sbjct 198 DNMIAQNLVELPMFSVYMSRDPNSPVGGELVFGGFDASRFSG--QLNWVPVTNQ--GYWQ 253
Query 118 LWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ L ++ ++ V FC+ C A++DTGTS
Sbjct 254 IQLDNIQINGEVV-FCSGG--CQAIVDTGTS 281
> xla:373563 pgc-A; pepsinogen C (EC:3.4.23.3); K01377 gastricsin
[EC:3.4.23.3]
Length=383
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 38/150 (25%), Positives = 73/150 (48%), Gaps = 8/150 (5%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + + G + + +Q V + G + F FDGI GL +P V +++++
Sbjct 131 GSLTGILGYDTVTIQNVAISQQEFGLSETEPGTNFVYAQFDGILGLAYPSIAVGGATTVM 190
Query 60 KQLVEAGAIKLAMFAIALP-LSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWEL 118
+ +++ + +F L S+ + GEV FGG +++Y I W P+ S+ W++
Sbjct 191 QGMMQQNLLNQPIFGFYLSGQSSQNGGEVAFGGVDQNYYTG--QIYWTPVTSE--TYWQI 246
Query 119 WLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ ++ G+C+ C A++DTGTS
Sbjct 247 GIQGFSINGQATGWCSQG--CQAIVDTGTS 274
> mmu:13034 Ctse, A430072O03Rik, C920004C08Rik, CE, CatE; cathepsin
E (EC:3.4.23.34); K01382 cathepsin E [EC:3.4.23.34]
Length=397
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 42/151 (27%), Positives = 75/151 (49%), Gaps = 10/151 (6%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQES-SSLI 59
G + + G + + ++ + + G V++ F FDGI GLG+P + +
Sbjct 143 GSLTGIIGADQVSVEGLTVDGQQFGESVKEPGQTFVNAEFDGILGLGYPSLAAGGVTPVF 202
Query 60 KQLVEAGAIKLAMFAIALPL--SASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ + L MF++ L S E+TFGG++ + ++ W P ++KQ+ W+
Sbjct 203 DNMMAQNLVALPMFSVYLSSDPQGGSGSELTFGGYDPSHFSG--SLNWIP-VTKQA-YWQ 258
Query 118 LWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ L + V D TV FC+ C A++DTGTS
Sbjct 259 IALDGIQVGD-TVMFCSEG--CQAIVDTGTS 286
> dre:114367 nots, LAP, MGC192423, ctsel, zgc:103608; nothepsin
Length=416
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/154 (27%), Positives = 68/154 (44%), Gaps = 6/154 (3%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESSS-LI 59
G++ + R+ L + V++ + G V + + F FDG+ GLG P +E S +
Sbjct 150 GHLLGVMARDELKVGSVRVQNQVFGEAVYEPGFSFVLAQFDGVLGLGFPQLAEEKGSPVF 209
Query 60 KQLVEAGAIKLAMFAIALPLSASS-AGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWEL 118
++E + +F+ L + S GE+ FG + I W P+ Q W++
Sbjct 210 DTMMEQNMLDQPVFSFYLTNNGSGFGGELVFGANDESRFLP--PINWIPV--TQKGYWQI 265
Query 119 WLWDVLVDDATVGFCTPQLPCIAVIDTGTSGDGG 152
L V V A C A++DTGTS GG
Sbjct 266 KLDAVKVQGALSFSDRSVQGCQAIVDTGTSLIGG 299
> hsa:1510 CTSE, CATE; cathepsin E (EC:3.4.23.34); K01382 cathepsin
E [EC:3.4.23.34]
Length=396
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 76/151 (50%), Gaps = 10/151 (6%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + + G + + ++ + + G V + F + FDGI GLG+P V + +
Sbjct 142 GSLSGIIGADQVSVEGLTVVGQQFGESVTEPGQTFVDAEFDGILGLGYPSLAVGGVTPVF 201
Query 60 KQLVEAGAIKLAMFAIALPLS--ASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ + L MF++ + + + E+ FGG++ + ++ W P ++KQ+ W+
Sbjct 202 DNMMAQNLVDLPMFSVYMSSNPEGGAGSELIFGGYDHSHFSG--SLNWVP-VTKQA-YWQ 257
Query 118 LWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ L ++ V TV FC+ C A++DTGTS
Sbjct 258 IALDNIQV-GGTVMFCSEG--CQAIVDTGTS 285
> xla:373564 pga4, pga-A; pepsinogen 4, group I (pepsinogen A);
K06002 pepsin A [EC:3.4.23.1]
Length=384
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/149 (23%), Positives = 65/149 (43%), Gaps = 8/149 (5%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + G + L + +++ + G + PFDGI GL P +++ +
Sbjct 136 GSMSGFLGYDTLQVGNIQISNQMFGLSESEPGSFLYYSPFDGILGLAFPSIASSQATPVF 195
Query 60 KQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELW 119
+ G I +F++ L + V FGG + Y ++ W PL ++ W++
Sbjct 196 DNMWSQGLIPQNLFSVYLSSDGQTGSYVLFGGVDNSYYSG--SLNWVPLTAE--TYWQIT 251
Query 120 LWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
L V ++ + C+ C A++DTGTS
Sbjct 252 LDSVSINGQVIA-CSQS--CQAIVDTGTS 277
> ath:AT1G11910 aspartyl protease family protein; K08245 phytepsin
[EC:3.4.23.40]
Length=506
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 59/112 (52%), Gaps = 9/112 (8%)
Query 40 FDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSA--SSAGEVTFGGFNRHY 96
FDGI GLG +V +++ + +++ G IK +F+ L +A GE+ FGG + ++
Sbjct 186 FDGILGLGFQEISVGKAAPVWYNMLKQGLIKEPVFSFWLNRNADEEEGGELVFGGVDPNH 245
Query 97 IQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ + P+ Q W+ + DVL+ A GFC + C A+ D+GTS
Sbjct 246 FKG--KHTYVPV--TQKGYWQFDMGDVLIGGAPTGFC--ESGCSAIADSGTS 291
> ath:AT1G69100 aspartic-type endopeptidase
Length=367
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 63/131 (48%), Gaps = 12/131 (9%)
Query 26 FVVRQAEYPFSELPFDGIAGLG-HPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSA 84
F+ R + F + FDG+ GLG Q S ++ + +V+ I +F++ L
Sbjct 136 FLARNPDTYFRSVKFDGVIGLGIKSSRAQGSVTVWENMVKQKLITKPIFSLYLRPHKGDG 195
Query 85 GE------VTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVD-DATVGFCTPQL 137
GE + FGGF+ KG + + P + +RW++ + + ++ + FC +
Sbjct 196 GEDPNGGQIMFGGFDPKQF-KGEHV-YVP-MKLSDDRWKIKMSKIYINGKPAINFCD-DV 251
Query 138 PCIAVIDTGTS 148
C A++D+G++
Sbjct 252 ECTAMVDSGST 262
> hsa:5222 PGA5, PGA3, PGA4; pepsinogen 5, group I (pepsinogen
A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 8/111 (7%)
Query 39 PFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYI 97
PFDGI GL +P + ++ + + G + +F++ L S V FGG + Y
Sbjct 178 PFDGILGLAYPSISSSGATPVFDNIWNQGLVSQDLFSVYLSADDKSGSVVIFGGIDSSYY 237
Query 98 QKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
++ W P+ + W++ + + ++ T+ C C A++DTGTS
Sbjct 238 TG--SLNWVPVTVE--GYWQITVDSITMNGETIA-CAEG--CQAIVDTGTS 281
> hsa:643847 PGA4, FLJ58952, FLJ77962, PGA3, PGA5; pepsinogen
4, group I (pepsinogen A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 50/115 (43%), Gaps = 16/115 (13%)
Query 39 PFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYI 97
PFDGI GL +P + ++ + + G + +F++ L S V FGG + Y
Sbjct 178 PFDGILGLAYPSISSSGATPVFDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSYY 237
Query 98 QKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG----FCTPQLPCIAVIDTGTS 148
++ W P+ + W + VD T+ C C A++DTGTS
Sbjct 238 TG--SLNWVPVTVEG-------YWQITVDSITMNGEAIACAEG--CQAIVDTGTS 281
> hsa:643834 PGA3, DKFZp666J2410, PGA4, PGA5; pepsinogen 3, group
I (pepsinogen A) (EC:3.4.23.1); K06002 pepsin A [EC:3.4.23.1]
Length=388
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 50/115 (43%), Gaps = 16/115 (13%)
Query 39 PFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYI 97
PFDGI GL +P + ++ + + G + +F++ L S V FGG + Y
Sbjct 178 PFDGILGLAYPSISSSGATPVFDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSYY 237
Query 98 QKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG----FCTPQLPCIAVIDTGTS 148
++ W P+ + W + VD T+ C C A++DTGTS
Sbjct 238 TG--SLNWVPVTVEG-------YWQITVDSITMNGEAIACAEG--CQAIVDTGTS 281
> mmu:13033 Ctsd, CD, CatD; cathepsin D (EC:3.4.23.5); K01379
cathepsin D [EC:3.4.23.5]
Length=410
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 56/137 (40%), Gaps = 14/137 (10%)
Query 17 VKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAI 75
+K+ G +Q F FDGI G+G+PH +V + L++ + +F+
Sbjct 170 IKVEKQIFGEATKQPGIVFVAAKFDGILGMGYPHISVNNVLPVFDNLMQQKLVDKNIFSF 229
Query 76 ALPL--SASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG-- 131
L GE+ GG + Y +++ + K W V +D VG
Sbjct 230 YLNRDPEGQPGGELMLGGTDSKYYHG--ELSYLNVTRKA-------YWQVHMDQLEVGNE 280
Query 132 FCTPQLPCIAVIDTGTS 148
+ C A++DTGTS
Sbjct 281 LTLCKGGCEAIVDTGTS 297
> dre:405786 ren; renin (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=395
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 59/118 (50%), Gaps = 10/118 (8%)
Query 34 PFSELPFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSAS--SAGEVTFG 90
PF FDG+ G+G+P+ + + + +++ +K +F++ + GE+ G
Sbjct 174 PFILAKFDGVLGMGYPNVAIDGITPVFDRIMSQHVLKENVFSVYYSRDPTHIPGGELVLG 233
Query 91 GFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
G + +Y G + + +K+ +WE+ + V V A + FC C AVIDTG+S
Sbjct 234 GTDPNY-HTG---PFHYINTKEQGKWEVIMKGVSV-GADILFCKDG--CTAVIDTGSS 284
> hsa:1509 CTSD, CLN10, CPSD, MGC2311; cathepsin D (EC:3.4.23.5);
K01379 cathepsin D [EC:3.4.23.5]
Length=412
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 57/137 (41%), Gaps = 14/137 (10%)
Query 17 VKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAI 75
VK+ G +Q F FDGI G+ +P +V + L++ + +F+
Sbjct 172 VKVERQVFGEATKQPGITFIAAKFDGILGMAYPRISVNNVLPVFDNLMQQKLVDQNIFSF 231
Query 76 ALPLS--ASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATV--G 131
L A GE+ GG + Y + ++++ + K W V +D V G
Sbjct 232 YLSRDPDAQPGGELMLGGTDSKYYKG--SLSYLNVTRKA-------YWQVHLDQVEVASG 282
Query 132 FCTPQLPCIAVIDTGTS 148
+ C A++DTGTS
Sbjct 283 LTLCKEGCEAIVDTGTS 299
> dre:336746 fa94d12, wu:fa94d12; zgc:63831; K01379 cathepsin
D [EC:3.4.23.5]
Length=412
Score = 35.8 bits (81), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 32/153 (20%), Positives = 62/153 (40%), Gaps = 14/153 (9%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + ++ ++L + + V+Q F+ FDG+ G+ +P +V + +
Sbjct 158 GSLSGFISQDTVNLAGLNVTGQQFAEAVKQPGIVFAVARFDGVLGMAYPAISVDRVTPVF 217
Query 60 KQLVEAGAIKLAMFA--IALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
+ A + +F+ I + GE+ GGF++ Y + N
Sbjct 218 DTAMAAKILPQNIFSFYINRDPAGDVGGELMLGGFDQQYFNGDLHYV---------NVTR 268
Query 118 LWLWDVLVDDATVG--FCTPQLPCIAVIDTGTS 148
W + +D+ VG + C A++DTGTS
Sbjct 269 KAYWQIKMDEVQVGSTLTLCKSGCQAIVDTGTS 301
> hsa:9476 NAPSA, KAP, Kdap, NAP1, NAPA, SNAPA; napsin A aspartic
peptidase; K08565 napsin-A [EC:3.4.23.-]
Length=420
Score = 35.4 bits (80), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 64/152 (42%), Gaps = 12/152 (7%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHP-HNVQESSSLI 59
G +D + + L + +K G + + F+ FDGI GLG P +V+ +
Sbjct 144 GRVDGILSEDKLTIGGIKGASVIFGEALWEPSLVFAFAHFDGILGLGFPILSVEGVRPPM 203
Query 60 KQLVEAGAIKLAMFAIALPLSASS--AGEVTFGGFN-RHYIQKGVAIAWFPLLSKQSNRW 116
LVE G + +F+ L GE+ GG + HYI + + P+ W
Sbjct 204 DVLVEQGLLDKPVFSFYLNRDPEEPDGGELVLGGSDPAHYIPP---LTFVPVTVPA--YW 258
Query 117 ELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
++ + V V C C A++DTGTS
Sbjct 259 QIHMERVKVGPGLT-LCAKG--CAAILDTGTS 287
> mmu:58803 Pga5, 1110035E17Rik, Pepf; pepsinogen 5, group I (EC:3.4.23.1);
K06002 pepsin A [EC:3.4.23.1]
Length=387
Score = 35.4 bits (80), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 51/116 (43%), Gaps = 8/116 (6%)
Query 40 FDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQ 98
FDGI GLG+P+ +Q + + L G I +FA L + GG + Y
Sbjct 177 FDGILGLGYPNLGLQGITPVFDNLWLQGLIPQNLFAFYLSSKDEKGSMLMLGGVDPSYYH 236
Query 99 KGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTSGDGGDR 154
+ W P+ + + W+L + D + + V C C ++DTGTS G R
Sbjct 237 G--ELHWVPV--SKPSYWQLAV-DSISMNGEVIACDGG--CQGIMDTGTSLLTGPR 285
> mmu:100044656 renin-1-like
Length=425
Score = 35.0 bits (79), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 57/120 (47%), Gaps = 15/120 (12%)
Query 34 PFSELPFDGIAGLGHP-HNVQESSSLIKQLVEAGAIKLAMFAIALPL-SASSAGEVTFGG 91
PF FDG+ G+G P V + + ++ G +K +F++ S GEV GG
Sbjct 205 PFMLAKFDGVLGMGFPAQAVGGVTPVFDHILSQGVLKEEVFSVYYNRGSHLLGGEVVLGG 264
Query 92 FNRHYIQKG---VAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ + Q V+I+ +++ W++ + V V +T+ C + C V+DTG+S
Sbjct 265 SDPQHYQGNFHYVSIS-------KTDSWQITMKGVSVGSSTL-LC--EEGCAVVVDTGSS 314
> mmu:19701 Ren1, D19352, Ren, Ren-1, Ren-A, Ren1c, Ren1d, Rn-1,
Rnr; renin 1 structural (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=402
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 57/120 (47%), Gaps = 15/120 (12%)
Query 34 PFSELPFDGIAGLGHP-HNVQESSSLIKQLVEAGAIKLAMFAIALPL-SASSAGEVTFGG 91
PF FDG+ G+G P V + + ++ G +K +F++ S GEV GG
Sbjct 182 PFMLAKFDGVLGMGFPAQAVGGVTPVFDHILSQGVLKEEVFSVYYNRGSHLLGGEVVLGG 241
Query 92 FNRHYIQKG---VAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ + Q V+I+ +++ W++ + V V +T+ C + C V+DTG+S
Sbjct 242 SDPQHYQGNFHYVSIS-------KTDSWQITMKGVSVGSSTL-LC--EEGCAVVVDTGSS 291
> pfa:PF14_0077 plasmepsin II; K06008 plasmepsin II [EC:3.4.23.39]
Length=453
Score = 35.0 bits (79), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 52/119 (43%), Gaps = 12/119 (10%)
Query 32 EYPFSELPFDGIAGLG-HPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFG 90
E ++ FDGI GLG ++ ++ +L I+ A+F LP+ G +T G
Sbjct 236 EPTYTASTFDGILGLGWKDLSIGSVDPIVVELKNQNKIENALFTFYLPVHDKHTGFLTIG 295
Query 91 GFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTSG 149
G + + PL ++ N +L+ W + +D + CI +D+GTS
Sbjct 296 GIEERFYEG-------PLTYEKLNH-DLY-WQITLDAHVGNIMLEKANCI--VDSGTSA 343
> ath:AT4G22050 aspartyl protease family protein
Length=354
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 4/61 (6%)
Query 35 FSELPFDGIAGLGHPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASS----AGEVTFG 90
F ++PFDGI GL + +S+ +V G I +F+I L ++S GEV FG
Sbjct 145 FKKMPFDGILGLRFTDPLNFGTSVWHSMVFQGKIAKNVFSIWLRRFSNSGEINGGEVVFG 204
Query 91 G 91
G
Sbjct 205 G 205
> xla:443721 ctsd; cathepsin D (EC:3.4.23.5); K01379 cathepsin
D [EC:3.4.23.5]
Length=399
Score = 34.3 bits (77), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 53/128 (41%), Gaps = 18/128 (14%)
Query 28 VRQAEYPFSELPFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSASS--A 84
V+Q F FDGI G+G+P +V + ++E + +F+ L + +
Sbjct 172 VKQPGITFVAAKFDGILGMGYPRISVDGVPPVFDDIMEQKLVDSNLFSFYLNRNPDTQPG 231
Query 85 GEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG----FCTPQLPCI 140
GE+ GG + Y + N W + +D +VG C + C
Sbjct 232 GELLLGGTDPTYYTGDFSYM---------NVTRKAYWQIRMDQLSVGDQLTLC--KGGCE 280
Query 141 AVIDTGTS 148
A++DTGTS
Sbjct 281 AIVDTGTS 288
> bbo:BBOV_IV009660 23.m06138; aspartyl protease
Length=435
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 37 ELPFDGIAGLGHPHNVQES--SSLIKQLVEAGAIKLAMFAIALPLSAS-SAGEVTFGGFN 93
+LP+DGI GLG + Q S S + Q ++ + F + + G VTFGG+
Sbjct 145 DLPWDGILGLGFITDDQISRGSKPLLQSIQDDELMYPNFRNQFAYYVTKNGGSVTFGGYK 204
Query 94 RHYIQK-GVAIAWFPLLSKQS 113
Y + G W P+ SK S
Sbjct 205 NEYKKSPGDVFQWAPVASKGS 225
> mmu:229697 Cym, Gm131; chymosin (EC:3.4.23.4); K01378 chymosin
[EC:3.4.23.4]
Length=379
Score = 33.9 bits (76), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 33/159 (20%), Positives = 66/159 (41%), Gaps = 16/159 (10%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESS-SLI 59
G ++ + + + + + +G ++ F+ PFDGI GL +P + S +
Sbjct 138 GRMEGFLAYDTVTVSDIVVSHQTVGLSTQEPGDIFTYSPFDGILGLAYPTFASKYSVPIF 197
Query 60 KQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELW 119
++ + +F++ + + + +T G ++ Y ++ W P+ +
Sbjct 198 DNMMNRHLVAQDLFSVYMSRNEQGS-MLTLGAIDQSYFIG--SLHWVPVTVQG------- 247
Query 120 LWDVLVDDATVG--FCTPQLPCIAVIDTGT---SGDGGD 153
W VD T+ Q C AV+DTGT +G G D
Sbjct 248 YWQFTVDRITINGEVVACQGGCPAVLDTGTALLTGPGRD 286
> cel:ZK384.3 hypothetical protein
Length=392
Score = 33.9 bits (76), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 60/148 (40%), Gaps = 17/148 (11%)
Query 8 GREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLIKQLVEAG 66
G + + + +K+ G V +Y F P DGI GL P +V + + ++ LV
Sbjct 139 GTDTVEIGGLKIQKQEFG-VANIVDYDFGTRPIDGIFGLAWPALSVDQVTPPMQNLVSQN 197
Query 67 AIKLAMFAIAL-----PLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLW 121
+ +F I S G +T+GG + I++ P+ SK W
Sbjct 198 QLDAPIFTIFFDKRDPDYYYPSNGLITYGGLDTKNCNAN--ISYVPVSSKT-------YW 248
Query 122 DVLVDDATVGFCTPQLPC-IAVIDTGTS 148
VD VG + A++DTG+S
Sbjct 249 QFKVDGFQVGTYNRTVNRDQAIMDTGSS 276
> mmu:19702 Ren2, Ren, Ren-2, Ren-B, Rn-2, Rnr; renin 2 tandem
duplication of Ren1 (EC:3.4.23.15); K01380 renin [EC:3.4.23.15]
Length=424
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 55/117 (47%), Gaps = 9/117 (7%)
Query 34 PFSELPFDGIAGLGHP-HNVQESSSLIKQLVEAGAIKLAMFAIALPLSAS-SAGEVTFGG 91
PF FDG+ G+G P V + + ++ G +K +F++ GEV GG
Sbjct 204 PFMLAQFDGVLGMGFPAQAVGGVTPVFDHILSQGVLKEKVFSVYYNRGPHLLGGEVVLGG 263
Query 92 FNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ + Q + LSK ++ W++ + V V +T+ C + C V+DTG+S
Sbjct 264 SDPEHYQGDFH---YVSLSK-TDSWQITMKGVSVGSSTL-LC--EEGCEVVVDTGSS 313
> pfa:PF14_0075 plasmepsin IV
Length=449
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 47/110 (42%), Gaps = 12/110 (10%)
Query 40 FDGIAGLG-HPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQ 98
FDGI GLG ++ ++ +L + I A+F LP+ G +T GG + +
Sbjct 241 FDGILGLGWKDLSIGSIDPVVVELKKQNKIDNALFTFYLPVHDKHVGYLTIGGIESDFYE 300
Query 99 KGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
PL ++ N W D+ D G Q AV+D+GTS
Sbjct 301 G-------PLTYEKLNHDLYWQIDL---DIHFGKYVMQ-KANAVVDSGTS 339
> xla:398994 napsa, MGC68767, kap, kdap, nap1, napa, snapa; napsin
A aspartic peptidase; K01379 cathepsin D [EC:3.4.23.5]
Length=392
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 32/151 (21%), Positives = 68/151 (45%), Gaps = 10/151 (6%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + ++ + + +++ + ++Q F FDGI G+G+P +V +
Sbjct 133 GSLSGFLSQDTVSIGSIEVANQTFAEAIKQPGIVFVFAHFDGILGMGYPDISVDGVVPVF 192
Query 60 KQLVEAGAIKLAMFAIALPLS--ASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
+++ ++ +F+ L A+ GE+ GG + +Y + L + W+
Sbjct 193 DNMMQQNLLEENVFSFYLSRDPMATVGGELILGGTDPNYYTGD----FHYLNVTRMAYWQ 248
Query 118 LWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+ +V V++ V C + C A++DTGTS
Sbjct 249 IKADEVRVNNQLV-LC--KGGCQAIVDTGTS 276
> mmu:16541 Napsa, KAP, Kdap, NAP1, SNAPA, pronapsin; napsin A
aspartic peptidase; K08565 napsin-A [EC:3.4.23.-]
Length=419
Score = 32.0 bits (71), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 51/117 (43%), Gaps = 20/117 (17%)
Query 40 FDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIALPLSA--SSAGEVTFGGFN-RH 95
FDGI GLG P V + +VE G ++ +F+ L + S GE+ GG + H
Sbjct 178 FDGILGLGFPTLAVGGVQPPLDAMVEQGLLEKPVFSFYLNRDSEGSDGGELVLGGSDPAH 237
Query 96 YIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVG----FCTPQLPCIAVIDTGTS 148
Y+ + + P+ W V ++ VG C C A++DTGTS
Sbjct 238 YVPP---LTFIPVTIPA-------YWQVHMESVKVGTGLSLCAQG--CSAILDTGTS 282
> cel:C15C8.3 hypothetical protein
Length=428
Score = 32.0 bits (71), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query 35 FSELPFDGIAGLGHPH-NVQESSSLIKQLVEAGAIKLAMFAIAL----PLSASSAGEVTF 89
F++ P DGI GLG P V + + + L+ G + F + L P S + G T
Sbjct 211 FAKQPEDGIIGLGWPALAVNQQTPPLFNLMNQGKLDQPYFVVYLANIGPTSQINGGAFTV 270
Query 90 GGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDV 123
GG + + V W PL ++ W+ L V
Sbjct 271 GGLDTTHCSSNV--DWVPLSTQ--TFWQFKLGGV 300
> hsa:5972 REN, FLJ10761, HNFJ2; renin (EC:3.4.23.15); K01380
renin [EC:3.4.23.15]
Length=406
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 29/129 (22%), Positives = 57/129 (44%), Gaps = 12/129 (9%)
Query 25 GFVVRQAEYPFSELPFDGIAGLGH-PHNVQESSSLIKQLVEAGAIKLAMFAIALPL---- 79
G V PF FDG+ G+G + + + ++ G +K +F+
Sbjct 175 GEVTEMPALPFMLAEFDGVVGMGFIEQAIGRVTPIFDNIISQGVLKEDVFSFYYNRDSEN 234
Query 80 SASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPC 139
S S G++ GG + + + + L+ ++ W++ + V V +T+ C + C
Sbjct 235 SQSLGGQIVLGGSDPQHYEGN--FHYINLI--KTGVWQIQMKGVSVGSSTL-LC--EDGC 287
Query 140 IAVIDTGTS 148
+A++DTG S
Sbjct 288 LALVDTGAS 296
> pfa:PF14_0076 plasmepsin I; K06007 plasmepsin I [EC:3.4.23.38]
Length=452
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 46/111 (41%), Gaps = 12/111 (10%)
Query 40 FDGIAGLG-HPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQ 98
FDGI GLG ++ ++ +L I+ A+F LP G +T GG + +
Sbjct 243 FDGIVGLGWKDLSIGSVDPVVVELKNQNKIEQAVFTFYLPFDDKHKGYLTIGGIEDRFYE 302
Query 99 KGVAIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTSG 149
L ++ N W D+ D G T + A++D+GTS
Sbjct 303 G-------QLTYEKLNHDLYWQVDL---DLHFGNLTVE-KATAIVDSGTSS 342
> dre:65225 ctsd, catD, fb93e11, fj17b09, wu:fb93e11, wu:fj17b09;
cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3.4.23.5]
Length=399
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 30/153 (19%), Positives = 63/153 (41%), Gaps = 14/153 (9%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + ++ + + + G ++Q F FDGI G+ +P +V +
Sbjct 141 GSLSGYLSQDTCTIGDIAVEKQIFGEAIKQPGVAFIAAKFDGILGMAYPRISVDGVPPVF 200
Query 60 KQLVEAGAIKLAMFAIALPLSASS--AGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ ++ +F+ L + + GE+ GG + Y + +S+Q+
Sbjct 201 DMMMSQKKVEKNVFSFYLNRNPDTQPGGELLLGGTDPKYYTGDFN---YVDISRQA---- 253
Query 118 LWLWDVLVDDATVG--FCTPQLPCIAVIDTGTS 148
W + +D ++G + C A++DTGTS
Sbjct 254 --YWQIHMDGMSIGSGLSLCKGGCEAIVDTGTS 284
> xla:443829 MGC82347 protein; K01379 cathepsin D [EC:3.4.23.5]
Length=401
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 32/153 (20%), Positives = 63/153 (41%), Gaps = 14/153 (9%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + ++ + + + + + G ++Q F FDGI G+ +P +V S +
Sbjct 147 GSLSGYLSKDTVTIGNLGIKEQLFGEAIKQPGVTFIAAKFDGILGMAYPIISVDGVSPVF 206
Query 60 KQLVEAGAIKLAMFAIALPLSASS--AGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ ++ +F+ L + + GE+ GG + Y F L+ +
Sbjct 207 DNIMAQKLVESNVFSFYLNRNPDTQPGGELLLGGTDPKYY-----TGDFHYLNVTRKAY- 260
Query 118 LWLWDVLVDDATVG--FCTPQLPCIAVIDTGTS 148
W + +D VG + C A++DTGTS
Sbjct 261 ---WQIHMDQLGVGDQLTLCKGGCEAIVDTGTS 290
> cel:Y39B6A.20 asp-1; ASpartyl Protease family member (asp-1)
Length=396
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 35/154 (22%), Positives = 60/154 (38%), Gaps = 16/154 (10%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G + G++VL+ + + G A+ F P DGI GLG P V + +
Sbjct 139 GSCNGYLGKDVLNFGGLTVQSQEFGVSTHLADV-FGYQPVDGILGLGWPALAVDQVVPPM 197
Query 60 KQLVEAGAIKLAMFAIALPLS-----ASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSN 114
+ L+ + +F + L + + G +T+G + K + + PL +K
Sbjct 198 QNLIAQKQLDAPLFTVWLDRNLQIAQGTPGGLITYGAIDTVNCAK--QVTYVPLSAKT-- 253
Query 115 RWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
W +D VG + + DTGTS
Sbjct 254 -----YWQFPLDAFAVGTYSETKKDQVISDTGTS 282
> hsa:6484 ST3GAL4, CGS23, FLJ11867, FLJ46764, NANTA3, SAT3, SIAT4,
SIAT4C, ST3GalIV, STZ; ST3 beta-galactoside alpha-2,3-sialyltransferase
4 (EC:2.4.99.4); K03494 beta-galactoside alpha-2,3-sialyltransferase
(sialyltransferase 4C) [EC:2.4.99.4]
Length=329
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 22 YAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESSSLIKQLVEAGAIK 69
Y + +Q + + ++ +AG GH NV + + IK+++E GAIK
Sbjct 279 YPDAYNKKQTIHYYEQITLKSMAGSGH--NVSQEALAIKRMLEMGAIK 324
> ath:AT1G44130 nucellin protein, putative
Length=405
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 17/107 (15%)
Query 42 GIAGLGHPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHYIQKGV 101
G+ GLG + L+ QLV AG L + LS+ G + FG + +
Sbjct 180 GVLGLG-----RGKIGLLTQLVSAG---LTRNVVGHCLSSKGGGFLFFG----DNLVPSI 227
Query 102 AIAWFPLLSKQSNRWELWLWDVLVDDATVGFCTPQLPCIAVIDTGTS 148
+AW PLLS Q N + D+L + G +L + DTG+S
Sbjct 228 GVAWTPLLS-QDNHYTTGPADLLFNGKPTGLKGLKL----IFDTGSS 269
> hsa:7272 TTK, CT96, ESK, FLJ38280, MPS1, MPS1L1, PYT; TTK protein
kinase (EC:2.7.12.1); K08866 serine/threonine-protein
kinase TTK/MPS1 [EC:2.7.12.1]
Length=856
Score = 29.6 bits (65), Expect = 4.1, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 51 NVQESSSLIKQLVEAGAIKLAMFAIAL 77
NV++S L+++ VE GA+ L M IAL
Sbjct 158 NVKKSKQLLQKAVERGAVPLEMLEIAL 184
> cpv:cgd1_3690 aspartyl (acid) protease
Length=792
Score = 29.3 bits (64), Expect = 4.6, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 27/55 (49%), Gaps = 2/55 (3%)
Query 42 GIAGLGHPHNVQESSSLIKQLVEAGAIKLAMFAIALPLSASSAGEVTFGGFNRHY 96
GI GL + + QE + L + + K A AI +S G ++FGG+N Y
Sbjct 173 GILGLAYTASSQERAPLFQTWTKRS--KYAKDAILSLCFSSEGGMISFGGYNSEY 225
> xla:398557 cathepsin D (EC:3.4.23.5); K01379 cathepsin D [EC:3.4.23.5]
Length=409
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 35/156 (22%), Positives = 62/156 (39%), Gaps = 20/156 (12%)
Query 1 GYIDSMTGREVLHLQKVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPH-NVQESSSLI 59
G I ++ + + + + G ++Q F FDGI G+ +P +V S
Sbjct 145 GSISGYLSKDTVTIGNLGYKEQIFGEAIKQPGVTFIAAKFDGILGMAYPIISVDGVSPCF 204
Query 60 KQLVEAGAIKLAMFAIALPLSASS--AGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWE 117
++ ++ +F+ L + + GE+ GG + Y F L+ +
Sbjct 205 DNIMAQKLVESNVFSFYLNRNPDTQPGGELLLGGTDPKYYTGD-----FHYLNVTRKAY- 258
Query 118 LWLWDVLVDDATVGFCTPQLP-----CIAVIDTGTS 148
W + +D VG QL C A++DTGTS
Sbjct 259 ---WQIHMDQLGVG---DQLTLCKGGCEAIVDTGTS 288
> cel:F59D6.2 hypothetical protein
Length=380
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 33/141 (23%), Positives = 55/141 (39%), Gaps = 20/141 (14%)
Query 16 KVKLHDYAIGFVVRQAEYPFSELPFDGIAGLGHPHNVQESSS-----LIKQLVEAGAIKL 70
K++ D+ IG + S + FDG+ GLG P +S L+ QL + +
Sbjct 145 KIQNQDFGIGTDATR----LSGVTFDGVLGLGWPATALNGTSTTMQNLLPQLDQ--PLFT 198
Query 71 AMFAIALPLSASSAGEVTFGGFNRHYIQKGVAIAWFPLLSKQSNRWELWLWDVLVDDATV 130
F + + + GE+TFG + + Q + S LW +D +
Sbjct 199 TYFTKSSVHNGTVGGEITFGAIDTTHCQSQINYVRLAYDS---------LWSYSIDGFSF 249
Query 131 GFCTPQLPCIAVIDTGTSGDG 151
G + A+ DT +S G
Sbjct 250 GNYSRNQTDTAIPDTTSSYTG 270
Lambda K H
0.321 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40