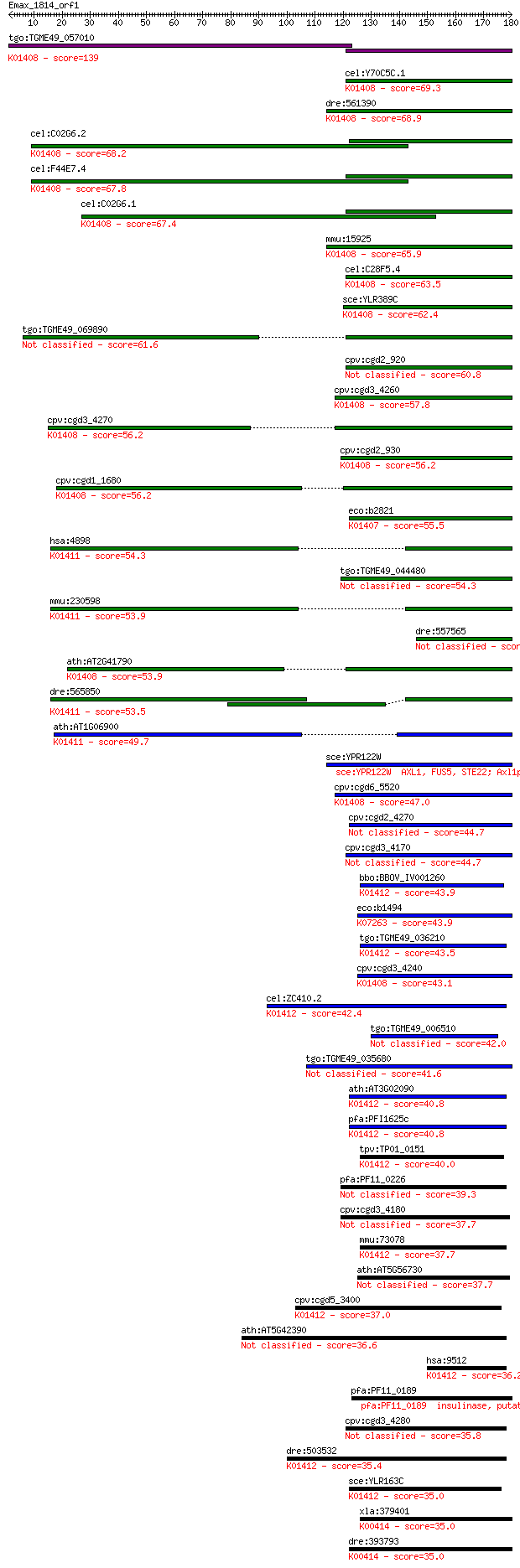
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_1814_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 139 4e-33
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 69.3 7e-12
dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzym... 68.9 8e-12
cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 68.2 1e-11
cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 67.8 2e-11
cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 67.4 2e-11
mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI50753... 65.9 6e-11
cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 63.5 3e-10
sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1... 62.4 8e-10
tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56) 61.6 1e-09
cpv:cgd2_920 peptidase'insulinase-like peptidase' 60.8 2e-09
cpv:cgd3_4260 peptidase'insulinase like peptidase' ; K01408 in... 57.8 2e-08
cpv:cgd3_4270 peptidase'insulinase like peptidase' ; K01408 in... 56.2 5e-08
cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 ins... 56.2 6e-08
cpv:cgd1_1680 insulinase like protease, signal peptide ; K0140... 56.2 6e-08
eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.... 55.5 1e-07
hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic co... 54.3 2e-07
tgo:TGME49_044480 insulin-degrading enzyme, putative (EC:3.4.2... 54.3 2e-07
mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nar... 53.9 2e-07
dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine diba... 53.9 3e-07
ath:AT2G41790 peptidase M16 family protein / insulinase family... 53.9 3e-07
dre:565850 fk24c07; wu:fk24c07; K01411 nardilysin [EC:3.4.24.61] 53.5 3e-07
ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptid... 49.7 5e-06
sce:YPR122W AXL1, FUS5, STE22; Axl1p (EC:3.4.24.-); K01422 [E... 47.8 2e-05
cpv:cgd6_5520 peptidase'insulinase like peptidase' ; K01408 in... 47.0 3e-05
cpv:cgd2_4270 secreted insulinase-like peptidase 44.7 2e-04
cpv:cgd3_4170 secreted insulinase like peptidase 44.7 2e-04
bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidas... 43.9 3e-04
eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted p... 43.9 3e-04
tgo:TGME49_036210 mitochondrial-processing peptidase beta subu... 43.5 4e-04
cpv:cgd3_4240 insulinase like peptidase ; K01408 insulysin [EC... 43.1 5e-04
cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta fa... 42.4 8e-04
tgo:TGME49_006510 peptidase M16 domain containing protein (EC:... 42.0 0.001
tgo:TGME49_035680 M16 family peptidase, putative (EC:4.1.1.70 ... 41.6 0.001
ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta... 40.8 0.002
pfa:PFI1625c organelle processing peptidase, putative; K01412 ... 40.8 0.003
tpv:TP01_0151 biquinol-cytochrome C reductase complex core pro... 40.0 0.004
pfa:PF11_0226 petidase, M16 family 39.3 0.008
cpv:cgd3_4180 secreted insulinase like peptidase 37.7 0.020
mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase... 37.7 0.021
ath:AT5G56730 peptidase M16 family protein / insulinase family... 37.7 0.023
cpv:cgd5_3400 mitochondrial processing peptidase beta subunit ... 37.0 0.032
ath:AT5G42390 metalloendopeptidase 36.6 0.043
hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase... 36.2 0.056
pfa:PF11_0189 insulinase, putative; K06972 35.8 0.084
cpv:cgd3_4280 secreted insulinase like peptidase, signal peptide 35.8 0.089
dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial process... 35.4 0.11
sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochond... 35.0 0.12
xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase ... 35.0 0.12
dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-c... 35.0 0.13
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 139 bits (351), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 60/122 (49%), Positives = 89/122 (72%), Gaps = 0/122 (0%)
Query 1 VGHELVSTMAQQSQIDFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLT 60
V + +S++++QS +DFHT+QP M+E ARLAHNLLTYEPYHV+AGDSLL+D D +
Sbjct 381 VDSKTISSISEQSLVDFHTSQPDPPAMNEVARLAHNLLTYEPYHVLAGDSLLVDPDAQFV 440
Query 61 NQLLQEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNA 120
NQLL +M+ AIIAF+DP F DSF+ +P+YG+++++ +LP+ + +T SP A
Sbjct 441 NQLLDKMTSDHAIIAFADPQFKRNNDSFDVEPFYGIEYKITNLPKEQRRRLETVTPSPGA 500
Query 121 FR 122
++
Sbjct 501 YK 502
Score = 102 bits (254), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 42/59 (71%), Positives = 51/59 (86%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
F H+QL NGM +A+HHP++ E ++VA NTGSLYDPED+PGLAHFLEHMLFLGTSK+P
Sbjct 33 FHHFQLPNGMQCLAIHHPKTTEGAYSVAVNTGSLYDPEDLPGLAHFLEHMLFLGTSKHP 91
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 69.3 bits (168), Expect = 7e-12, Method: Composition-based stats.
Identities = 29/59 (49%), Positives = 42/59 (71%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V P ++++ A+ N G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 26 YRGLELTNGIRVLLVSDPTTDKSAAALDVNVGHLMDPWELPGLAHFCEHMLFLGTAKYP 84
> dre:561390 ide, MGC162603, zgc:162603; insulin-degrading enzyme
(EC:3.4.24.56); K01408 insulysin [EC:3.4.24.56]
Length=978
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 32/69 (46%), Positives = 45/69 (65%), Gaps = 3/69 (4%)
Query 114 LTASPNAFRHY---QLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 170
+ SP R Y + +NG+ AI + P ++++ A+ + GSL DPE++ GLAHF EHM
Sbjct 13 IIRSPEDKREYRGLEFTNGLKAILISDPTTDKSSAALDVHMGSLSDPENISGLAHFCEHM 72
Query 171 LFLGTSKYP 179
LFLGT KYP
Sbjct 73 LFLGTEKYP 81
> cel:C02G6.2 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=816
Score = 68.2 bits (165), Expect = 1e-11, Method: Composition-based stats.
Identities = 29/58 (50%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
R +L+NG+ + V P ++++ ++A G L DP ++PGLAHF EHMLFLGTSKYP
Sbjct 27 RGLELTNGLRVLLVSDPTTDKSAVSLAVKAGHLMDPWELPGLAHFCEHMLFLGTSKYP 84
Score = 40.8 bits (94), Expect = 0.003, Method: Composition-based stats.
Identities = 32/135 (23%), Positives = 59/135 (43%), Gaps = 17/135 (12%)
Query 9 MAQQSQIDFHTTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLLQEM 67
+A+ S I+F + + A ++A NL Y P+ H+++ LL +P +LL +
Sbjct 377 LAELSAIEFRF-KDREPLTKNAIKVARNL-QYIPFEHILSSRYLLTKYNPERIKELLSTL 434
Query 68 SPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLS 127
+PS ++ F + + +P YG + +V D+ SP + Y+ +
Sbjct 435 TPSNMLVRVVSKKFKEQ-EGNTNEPVYGTEMKVTDI-------------SPEKMKKYENA 480
Query 128 NGMHAIAVHHPRSNE 142
A+H P NE
Sbjct 481 LKTSHHALHLPEKNE 495
> cel:F44E7.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=1051
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 85 YRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 143
Score = 34.3 bits (77), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 32/138 (23%), Positives = 57/138 (41%), Gaps = 23/138 (16%)
Query 9 MAQQSQIDFH---TTQPSSSIMDEAARLAHNLLTYEPY-HVVAGDSLLIDADPRLTNQLL 64
+A+ S + F QP + ++ AA L Y P+ H+++ LL +P +LL
Sbjct 436 LAELSAVKFRFKDKEQPMTMAINVAASLQ-----YIPFEHILSSRYLLTKYEPERIKELL 490
Query 65 QEMSPSKAIIAFSDPDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHY 124
+SP+ + F + + +P YG + +V D+ SP + Y
Sbjct 491 SMLSPANMQVRVVSQKFKGQ-EGNTNEPVYGTEMKVTDI-------------SPETMKKY 536
Query 125 QLSNGMHAIAVHHPRSNE 142
+ + A+H P NE
Sbjct 537 ENALKTSHHALHLPEKNE 554
> cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=980
Score = 67.4 bits (163), Expect = 2e-11, Method: Composition-based stats.
Identities = 28/59 (47%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V P ++++ A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 26 YRGLELTNGIRVLLVSDPTTDKSAAALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 84
Score = 37.4 bits (85), Expect = 0.025, Method: Composition-based stats.
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 17/126 (13%)
Query 27 MDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDFTSKVD 86
M A +A +L H+++ LL +P +LL ++PS ++ F + +
Sbjct 367 MKMAINIAASLQYIPIEHILSSRYLLTKYEPERIKELLSTLTPSNMLVRVVSQKFKEQ-E 425
Query 87 SFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFA 146
+P YG + +V D+ SP + Y+ + A+H P NE
Sbjct 426 GNTNEPVYGTEMKVTDI-------------SPEKMKKYENALKTSHHALHLPEKNE---Y 469
Query 147 VAANTG 152
+A N G
Sbjct 470 IATNFG 475
> mmu:15925 Ide, 1300012G03Rik, 4833415K22Rik, AA675336, AI507533;
insulin degrading enzyme (EC:3.4.24.56); K01408 insulysin
[EC:3.4.24.56]
Length=1019
Score = 65.9 bits (159), Expect = 6e-11, Method: Composition-based stats.
Identities = 30/69 (43%), Positives = 45/69 (65%), Gaps = 3/69 (4%)
Query 114 LTASPNAFRHY---QLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHM 170
+ SP R Y +L+NG+ + + P ++++ A+ + GSL DP ++PGL+HF EHM
Sbjct 54 IVKSPEDKREYRGLELANGIKVLLISDPTTDKSSAALDVHIGSLSDPPNIPGLSHFCEHM 113
Query 171 LFLGTSKYP 179
LFLGT KYP
Sbjct 114 LFLGTKKYP 122
> cel:C28F5.4 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=856
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 27/59 (45%), Positives = 39/59 (66%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L+NG+ + V ++ + A+ G L DP ++PGLAHF EHMLFLGT+KYP
Sbjct 27 YRGLELTNGLRVLLVSDSKTRVSAVALDVKVGHLMDPWELPGLAHFCEHMLFLGTAKYP 85
> sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1p,
in N-terminal processing of pro-A-factor to the mature
form; member of the insulin-degrading enzyme family (EC:3.4.24.-);
K01408 insulysin [EC:3.4.24.56]
Length=1027
Score = 62.4 bits (150), Expect = 8e-10, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 43/60 (71%), Gaps = 0/60 (0%)
Query 120 AFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++R +L N + A+ + P++++A ++ N G+ DP+++PGLAHF EH+LF+G+ K+P
Sbjct 73 SYRFIELPNKLKALLIQDPKADKAAASLDVNIGAFEDPKNLPGLAHFCEHLLFMGSEKFP 132
> tgo:TGME49_069890 M16 family peptidase, putative (EC:3.4.24.56)
Length=941
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R+ +L N + A+ V P +EA ++ GS+ DP +PGLAHF EHMLF G+ ++P
Sbjct 35 YRYIELPNELRALLVSDPECDEAAASMRVGVGSMSDPPKIPGLAHFTEHMLFQGSKRFP 93
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 36/86 (41%), Gaps = 4/86 (4%)
Query 6 VSTMAQQSQIDF-HTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLL 64
V+ MA+ Q+ F P + R L Y P V+AGD L+ DP + Q +
Sbjct 387 VTEMAKIRQLGFAFADMPDPYAL--TVRAVEGLNYYTPEEVIAGDRLIYHFDPDIIQQYV 444
Query 65 QE-MSPSKAIIAFSDPDFTSKVDSFE 89
Q+ + P + D + VD E
Sbjct 445 QKFLVPDNVRLFIFDKKLAADVDREE 470
> cpv:cgd2_920 peptidase'insulinase-like peptidase'
Length=1028
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R +L NG+ A V + +G + G++Y P+++ GLAHFLEHMLF GT KYP
Sbjct 27 YRALELKNGLTAFLVSDKETKTSGCCLTVYIGAMYSPKNLNGLAHFLEHMLFCGTKKYP 85
> cpv:cgd3_4260 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=1172
Score = 57.8 bits (138), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 117 SPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
S FR+ +L N + V H + + +A GS +P+ PGLAH+LEH+LF+ T
Sbjct 66 SDKQFRYIKLKNELEVFLVSHNDTKVSSANIAVKVGSYMEPDSFPGLAHYLEHLLFINTE 125
Query 177 KYP 179
KYP
Sbjct 126 KYP 128
> cpv:cgd3_4270 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=1176
Score = 56.2 bits (134), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 117 SPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
S FR+ +L N + V H + + + GS +P+ PGLAH+LEH+LF+ T
Sbjct 70 SDKQFRYIKLKNELEVFLVSHNDTKVSSANIVVKVGSYMEPDSFPGLAHYLEHLLFINTE 129
Query 177 KYP 179
KYP
Sbjct 130 KYP 132
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 17/74 (22%), Positives = 33/74 (44%), Gaps = 3/74 (4%)
Query 15 IDFHTTQPSSSIMDEAARLAHNLLTYE--PYHVVAGDSLLIDADPRLTNQLLQEMSPSKA 72
I F P+S+ D+ + + YE P V+ D + + DP + ++ +++P
Sbjct 437 ISFKYEDPTST-FDQCKEIVTYYIQYECKPEDVLYSDYYMDEFDPNIYKEINSQLTPENL 495
Query 73 IIAFSDPDFTSKVD 86
II PD ++
Sbjct 496 IITLERPDIEDLIN 509
> cpv:cgd2_930 peptidase'insulinase-like peptidase' ; K01408 insulysin
[EC:3.4.24.56]
Length=1013
Score = 56.2 bits (134), Expect = 6e-08, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 119 NAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
N +R L N + + V ++ +G +++ G DPE++ GLAHFLEHMLFLG++++
Sbjct 23 NKYRALVLKNNLRVLLVQDENTDISGASMSVFVGCQQDPEELNGLAHFLEHMLFLGSARH 82
Query 179 P 179
P
Sbjct 83 P 83
> cpv:cgd1_1680 insulinase like protease, signal peptide ; K01408
insulysin [EC:3.4.24.56]
Length=1033
Score = 56.2 bits (134), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 120 AFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++++ L NG+ A+ + S +AGF V GS +P GL H +EH+LFLGT KYP
Sbjct 47 SYKNITLENGITALLIEDKFSEKAGFTVGIKVGSFNNPVYALGLFHLIEHVLFLGTKKYP 106
Score = 33.5 bits (75), Expect = 0.36, Method: Composition-based stats.
Identities = 21/88 (23%), Positives = 41/88 (46%), Gaps = 6/88 (6%)
Query 18 HTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFS 77
+T SS M + + ++ Y + ++GD L+ D D L ++L +SP + S
Sbjct 416 NTEMLESSPMHSTSEICSKMIQYGVHAALSGDILIEDVDENLIYEILNAISPFDTLFLVS 475
Query 78 DP-DFTSKVDSFETDPYYGVQFRVLDLP 104
D +F+ + F + V+ + D+P
Sbjct 476 DEQEFSGTYEKF-----FHVKHAIEDIP 498
> eco:b2821 ptrA, ECK2817, JW2789, ptr; protease III (EC:3.4.24.55);
K01407 protease III [EC:3.4.24.55]
Length=962
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Query 122 RHYQ---LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
R YQ L NGM + V P++ ++ A+ GSL DPE GLAH+LEHM +G+ KY
Sbjct 42 RQYQAIRLDNGMVVLLVSDPQAVKSLSALVVPVGSLEDPEAYQGLAHYLEHMSLMGSKKY 101
Query 179 P 179
P
Sbjct 102 P 102
> hsa:4898 NRD1, hNRD1, hNRD2; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61); K01411 nardilysin [EC:3.4.24.61]
Length=1151
Score = 54.3 bits (129), Expect = 2e-07, Method: Composition-based stats.
Identities = 21/38 (55%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 142 EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++ A+ GS DP+D+PGLAHFLEHM+F+G+ KYP
Sbjct 210 QSAAALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYP 247
Score = 32.3 bits (72), Expect = 0.80, Method: Composition-based stats.
Identities = 20/88 (22%), Positives = 39/88 (44%), Gaps = 3/88 (3%)
Query 16 DFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIA 75
+FH Q + ++ + N+ Y ++ GD LL + P + + L ++ P KA +
Sbjct 552 EFHY-QEQTDPVEYVENMCENMQLYPLQDILTGDQLLFEYKPEVIGEALNQLVPQKANLV 610
Query 76 FSDPDFTSKVDSFETDPYYGVQFRVLDL 103
K D E ++G Q+ + D+
Sbjct 611 LLSGANEGKCDLKEK--WFGTQYSIEDI 636
> tgo:TGME49_044480 insulin-degrading enzyme, putative (EC:3.4.24.56)
Length=299
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 119 NAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
++R +L N + V P ++ A A+ N GS +DP V GLAHF EHMLFLGT K+
Sbjct 17 RSYRFVKLPNHLSVWLVSDPAADLASAALDINVGSYFDPPPVEGLAHFCEHMLFLGTEKF 76
Query 179 P 179
P
Sbjct 77 P 77
> mmu:230598 Nrd1, 2600011I06Rik, AI875733, MGC25477, NRD-C; nardilysin,
N-arginine dibasic convertase, NRD convertase 1 (EC:3.4.24.61);
K01411 nardilysin [EC:3.4.24.61]
Length=1161
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 21/38 (55%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 142 EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++ A+ GS DP+D+PGLAHFLEHM+F+G+ KYP
Sbjct 221 QSAAALCVGVGSFADPDDLPGLAHFLEHMVFMGSLKYP 258
Score = 29.6 bits (65), Expect = 5.4, Method: Composition-based stats.
Identities = 19/88 (21%), Positives = 38/88 (43%), Gaps = 3/88 (3%)
Query 16 DFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIA 75
+FH Q + ++ + N+ Y + GD LL + P + + L ++ P KA +
Sbjct 563 EFHY-QEQTDPVEYVENMCENMQLYPRQDFLTGDQLLFEYKPEVIAEALNQLVPQKANLV 621
Query 76 FSDPDFTSKVDSFETDPYYGVQFRVLDL 103
+ D E ++G Q+ + D+
Sbjct 622 LLSGANEGRCDLKEK--WFGTQYSIEDI 647
> dre:557565 nrd1, si:dkey-171o17.4; nardilysin (N-arginine dibasic
convertase) (EC:3.4.24.61)
Length=1061
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/34 (61%), Positives = 27/34 (79%), Gaps = 0/34 (0%)
Query 146 AVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
A+ + GS DP D+PGLAHFLEHM+F+G+ KYP
Sbjct 118 ALCISVGSFSDPADLPGLAHFLEHMVFMGSEKYP 151
> ath:AT2G41790 peptidase M16 family protein / insulinase family
protein; K01408 insulysin [EC:3.4.24.56]
Length=970
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R L N + + + P +++ +++ + GS DP+ + GLAHFLEHMLF + KYP
Sbjct 25 YRMIVLKNLLQVLLISDPDTDKCAASMSVSVGSFSDPQGLEGLAHFLEHMLFYASEKYP 83
Score = 32.3 bits (72), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 35/77 (45%), Gaps = 6/77 (7%)
Query 22 PSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAFSDPDF 81
P S I+D +A N+ Y + G SL +P + +++ E+SPS I + F
Sbjct 391 PMSYIVD----IASNMQIYPTKDWLVGSSLPTKFNPAIVQKVVDELSPSNFRIFWESQKF 446
Query 82 TSKVDSFETDPYYGVQF 98
+ D E P+Y +
Sbjct 447 EGQTDKAE--PWYNTAY 461
> dre:565850 fk24c07; wu:fk24c07; K01411 nardilysin [EC:3.4.24.61]
Length=1091
Score = 53.5 bits (127), Expect = 3e-07, Method: Composition-based stats.
Identities = 21/38 (55%), Positives = 28/38 (73%), Gaps = 0/38 (0%)
Query 142 EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++ A+ GS DP D+PGLAHFLEHM+F+G+ KYP
Sbjct 145 QSAAALCIGVGSFSDPNDLPGLAHFLEHMVFMGSEKYP 182
Score = 32.3 bits (72), Expect = 0.98, Method: Composition-based stats.
Identities = 19/91 (20%), Positives = 40/91 (43%), Gaps = 3/91 (3%)
Query 16 DFHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIA 75
+FH + + I + + N+ + + GD L+ + P + + L ++P KA +
Sbjct 487 EFHYQEQTDPI-EYVEDICENMQLFPKEDFLTGDQLMFEFKPEVISAALNLLTPEKANLL 545
Query 76 FSDPDFTSKVDSFETDPYYGVQFRVLDLPQH 106
P+ + E ++G Q+ D+ QH
Sbjct 546 LLSPEHEGQCPLREK--WFGTQYSTEDIEQH 574
Score = 31.2 bits (69), Expect = 2.2, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 4/56 (7%)
Query 79 PDFTSKVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIA 134
PDF+ K++ FET P + +RV+ + + H A+ + A P +HY++ +H IA
Sbjct 346 PDFSDKLNPFET-PAFNKLYRVVPVRKVH--ALTITWALPPQEKHYRVKP-LHYIA 397
> ath:AT1G06900 catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01411 nardilysin [EC:3.4.24.61]
Length=1024
Score = 49.7 bits (117), Expect = 5e-06, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 139 RSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
++ +A A+ + GS DP + GLAHFLEHMLF+G++++P
Sbjct 103 QTKKAAAAMCVSMGSFLDPPEAQGLAHFLEHMLFMGSTEFP 143
Score = 43.5 bits (101), Expect = 4e-04, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 41/88 (46%), Gaps = 5/88 (5%)
Query 17 FHTTQPSSSIMDEAARLAHNLLTYEPYHVVAGDSLLIDADPRLTNQLLQEMSPSKAIIAF 76
F QP+ D AA L+ N+L Y HV+ GD + DP+L L+ +P I
Sbjct 447 FAEEQPAD---DYAAELSENMLAYPVEHVIYGDYVYQTWDPKLIEDLMGFFTPQNMRIDV 503
Query 77 SDPDFTSKVDSFETDPYYGVQFRVLDLP 104
S + F+ +P++G + D+P
Sbjct 504 VSKSIKS--EEFQQEPWFGSSYIEEDVP 529
> sce:YPR122W AXL1, FUS5, STE22; Axl1p (EC:3.4.24.-); K01422
[EC:3.4.99.-]
Length=1208
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 114 LTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLF- 172
L+ S + +L NG+ A+ + P + ++ TGS DP+D+ GLAH EHM+
Sbjct 17 LSYSNRTHKVCKLPNGILALIISDPTDTSSSCSLTVCTGSHNDPKDIAGLAHLCEHMILS 76
Query 173 LGTSKYP 179
G+ KYP
Sbjct 77 AGSKKYP 83
> cpv:cgd6_5520 peptidase'insulinase like peptidase' ; K01408
insulysin [EC:3.4.24.56]
Length=570
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Query 117 SPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
+ N +R +L+N + + P + G + GS DPE +PGLAH L+ LF+ T
Sbjct 60 TENKYRFIKLNNELDVFLISRPGKHTYG-TLHIQVGSHNDPEYIPGLAHLLKQSLFINTK 118
Query 177 KYP 179
KYP
Sbjct 119 KYP 121
> cpv:cgd2_4270 secreted insulinase-like peptidase
Length=1257
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 32/58 (55%), Gaps = 0/58 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+ Y S G+ + V E+ F+ G DP+++ GLAH +EH++FLG+ + P
Sbjct 94 KAYTTSKGLKTLLVSDNTMLESAFSFGIGCGYYQDPDNLAGLAHLMEHVVFLGSQENP 151
> cpv:cgd3_4170 secreted insulinase like peptidase
Length=1289
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+R+ +L NG+ A V + ++ A+ + G LYDP + GL++ +++ L L + +YP
Sbjct 68 YRYIKLKNGLKAFLVSKEDAEKSEVAILVDVGFLYDPPKIIGLSNLVQYSLLLASYQYP 126
> bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidase
beta subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=514
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
L NG+ +V P N V ++GS ++ ++ G AHFLEHM+F GT
Sbjct 71 LKNGLRVASVWMP-GNSTTVGVWIDSGSRFETKETNGAAHFLEHMIFKGTK 120
> eco:b1494 pqqL, ECK1488, JW1489, pqqE, pqqM, yddC; predicted
peptidase; K07263 zinc protease [EC:3.4.24.-]
Length=931
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 37/57 (64%), Gaps = 3/57 (5%)
Query 125 QLSNGM-HAIAVH-HPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
QL NG+ + I H HP+ ++ + +TGSL + ++ G+AHF+EHM+F GT +P
Sbjct 39 QLDNGLRYMIYPHAHPK-DQVNLWLQIHTGSLQEEDNELGVAHFVEHMMFNGTKTWP 94
> tgo:TGME49_036210 mitochondrial-processing peptidase beta subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=524
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 27/52 (51%), Gaps = 0/52 (0%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
L NG+ P A V ++GS YD ++ G AHFLEHM F GT +
Sbjct 74 LPNGIRVATQRLPFHQTATVGVWIDSGSRYDTKETNGAAHFLEHMTFKGTKR 125
> cpv:cgd3_4240 insulinase like peptidase ; K01408 insulysin [EC:3.4.24.56]
Length=1113
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 0/55 (0%)
Query 125 QLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+L G+ + + + GS + ++ GLAHFLEH +FLGT K+P
Sbjct 36 RLKTGLEVFLISSEKLTSTSVNLVVKVGSANEGSEIDGLAHFLEHSVFLGTEKFP 90
> cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta
family member (mppb-1); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=458
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 39/86 (45%), Gaps = 11/86 (12%)
Query 93 YYGVQFRVLDL-PQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANT 151
Y Q R+ + P+ V ++T PN FR +A + + A V +
Sbjct 10 YQTSQRRIAQVQPKSVFVPETIVTTLPNGFR----------VATENTGGSTATIGVFIDA 59
Query 152 GSLYDPEDVPGLAHFLEHMLFLGTSK 177
GS Y+ E G AHFLEHM F GT +
Sbjct 60 GSRYENEKNNGTAHFLEHMAFKGTPR 85
> tgo:TGME49_006510 peptidase M16 domain containing protein (EC:4.1.1.70
3.4.24.13 3.4.24.56 3.4.21.10 3.4.24.35 3.2.1.91)
Length=2435
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 130 MHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLG 174
+ +A+ FAV+ G +DP +PG+AH LEH++FLG
Sbjct 224 LEGVAIADQEEAVGSFAVSVGCGFFHDPPAIPGVAHQLEHLIFLG 268
> tgo:TGME49_035680 M16 family peptidase, putative (EC:4.1.1.70
3.4.24.64)
Length=1692
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 43/88 (48%), Gaps = 25/88 (28%)
Query 107 HAVAMAVLTASPNAF----RHYQLSNGM-----------HAIAVHHPRSNEAGFAVAANT 151
+ A+ ++A+P AF R +L NG+ H IA H + +
Sbjct 440 YRAALQSVSATPLAFWPALRLGRLRNGLEYRILQHAFPAHKIAAH----------LVVHA 489
Query 152 GSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
GS+++ E+ GLAH LEH +F GT K+P
Sbjct 490 GSVHEEENEQGLAHLLEHCVFQGTRKFP 517
> ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta
subunit, putative; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=531
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
R L NG+ + + A V + GS ++ ++ G AHFLEHM+F GT +
Sbjct 98 RVTTLPNGLRVATESNLSAKTATVGVWIDAGSRFESDETNGTAHFLEHMIFKGTDR 153
> pfa:PFI1625c organelle processing peptidase, putative; K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=484
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
R +LSN + +A H + ++GS Y+ + G+AHFLEHM+F GT K
Sbjct 43 RVTELSNKL-KVATVHTNCEIPTIGLWISSGSKYENKKNNGVAHFLEHMIFKGTKK 97
> tpv:TP01_0151 biquinol-cytochrome C reductase complex core protein
I (EC:1.10.2.2); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=518
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTS 176
L NG+ V P S+ + V ++GS ++ + G AHFLEHM+F GT
Sbjct 77 LKNGLRVATVWMPGSS-STVGVWIDSGSRFETPETNGSAHFLEHMIFKGTK 126
> pfa:PF11_0226 petidase, M16 family
Length=2024
Score = 39.3 bits (90), Expect = 0.008, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query 119 NAFRHYQL-SNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
N + +++L SN + + + + S + GF+++ N G D ++PG+++ L H +F + K
Sbjct 676 NEYEYFKLKSNELKVLGIINKYSPKGGFSISVNCGGYDDFREIPGISNLLRHAIFYKSEK 735
> cpv:cgd3_4180 secreted insulinase like peptidase
Length=1215
Score = 37.7 bits (86), Expect = 0.020, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 119 NAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
+ ++ +L N M V + + ++ GS+ DPED+PGL ++ L LGT ++
Sbjct 80 STYKFMKLQNQMSVFLVSNNNFEYSIITLSVGVGSVMDPEDLPGLVSLVQESLCLGTYRF 139
> mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 37.7 bits (86), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
L NG+ +A + + + + GS Y+ E G AHFLEHM F GT K
Sbjct 63 LENGLR-VASENSGLSTCTVGLWIDAGSRYENEKNNGTAHFLEHMAFKGTKK 113
> ath:AT5G56730 peptidase M16 family protein / insulinase family
protein
Length=956
Score = 37.7 bits (86), Expect = 0.023, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 9/59 (15%)
Query 125 QLSNGMHAIAVHHPRSN-----EAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKY 178
+L NG+ +++ R N A A+A GS+ + ED G+AH +EH+ F T++Y
Sbjct 44 RLDNGL----IYYVRRNSKPRMRAALALAVKVGSVLEEEDQRGVAHIVEHLAFSATTRY 98
> cpv:cgd5_3400 mitochondrial processing peptidase beta subunit
; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=375
Score = 37.0 bits (84), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 37/78 (47%), Gaps = 5/78 (6%)
Query 103 LPQHHAVAMAVLTASPNA--FRHYQLSNGMHAIAVHHPRS---NEAGFAVAANTGSLYDP 157
LP +A + L+ N + +LSNGM + N F + ++GS +
Sbjct 21 LPTRYARFNSTLSFKRNDPDLKISKLSNGMRVATMKFGIDSIPNSLTFGLWVDSGSRNED 80
Query 158 EDVPGLAHFLEHMLFLGT 175
G+AHFLEH++F GT
Sbjct 81 PGKNGIAHFLEHLIFKGT 98
> ath:AT5G42390 metalloendopeptidase
Length=1265
Score = 36.6 bits (83), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 46/97 (47%), Gaps = 22/97 (22%)
Query 84 KVDSFETDPYYGVQFRVLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHH---PRS 140
++DS E + + G + LP H P R QL NG+ + + + P
Sbjct 175 EIDSAELEAFLGCE-----LPSH-----------PKLHRG-QLKNGLRYLILPNKVPPNR 217
Query 141 NEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
EA V + GS+ + ED G+AH +EH+ FLG+ K
Sbjct 218 FEAHMEV--HVGSIDEEEDEQGIAHMIEHVAFLGSKK 252
> hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 36.2 bits (82), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 150 NTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
+ GS Y+ E G AHFLEHM F GT K
Sbjct 86 DAGSRYENEKNNGTAHFLEHMAFKGTKK 113
> pfa:PF11_0189 insulinase, putative; K06972
Length=1488
Score = 35.8 bits (81), Expect = 0.084, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 5/57 (8%)
Query 123 HYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
+Y L +G+ I GF L + E+ GL H LEH++FLG+ KYP
Sbjct 21 YYSLDSGLRIILNKTKSPKIYGFFTL-----LTEAENDEGLPHTLEHLIFLGSHKYP 72
> cpv:cgd3_4280 secreted insulinase like peptidase, signal peptide
Length=1244
Score = 35.8 bits (81), Expect = 0.089, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 121 FRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSK 177
+R+ +LSN + V+ + + ++ + G DPE++PGL+ +L + L G+ K
Sbjct 113 YRYLRLSNSLKVFMVYDKTTEISFGSMNLDFGFASDPENIPGLSRYLLYTLLFGSLK 169
> dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=470
Score = 35.4 bits (80), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 34/78 (43%), Gaps = 16/78 (20%)
Query 100 VLDLPQHHAVAMAVLTASPNAFRHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPED 159
VL++P+ LT N R +G+ V G + A GS Y+ E
Sbjct 38 VLNVPETK------LTTLDNGLRVASEDSGLSTCTV--------GLWIDA--GSRYENEH 81
Query 160 VPGLAHFLEHMLFLGTSK 177
G AHFLEHM F GT K
Sbjct 82 NNGTAHFLEHMAFKGTRK 99
> sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=462
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGT 175
R +L NG+ + P ++ A + + GS + G AHFLEH+ F GT
Sbjct 27 RTSKLPNGLTIATEYIPNTSSATVGIFVDAGSRAENVKNNGTAHFLEHLAFKGT 80
> xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase
core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=478
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 1/54 (1%)
Query 126 LSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
L NG+ +A V GS Y+ + G +FLEH+ F GT K P
Sbjct 51 LDNGLR-VASEESSQATCTVGVWIGAGSRYESDKNNGAGYFLEHLAFKGTKKRP 103
> dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414
ubiquinol-cytochrome c reductase core subunit 1 [EC:1.10.2.2]
Length=474
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 25/58 (43%), Gaps = 1/58 (1%)
Query 122 RHYQLSNGMHAIAVHHPRSNEAGFAVAANTGSLYDPEDVPGLAHFLEHMLFLGTSKYP 179
R L NG+ IA + GS ++ E G FLEHM F GT K+P
Sbjct 43 RLTTLDNGLR-IASEETNQPTCTVGLWIGCGSRFETEKNNGAGFFLEHMAFKGTKKHP 99
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4795148792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40