bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
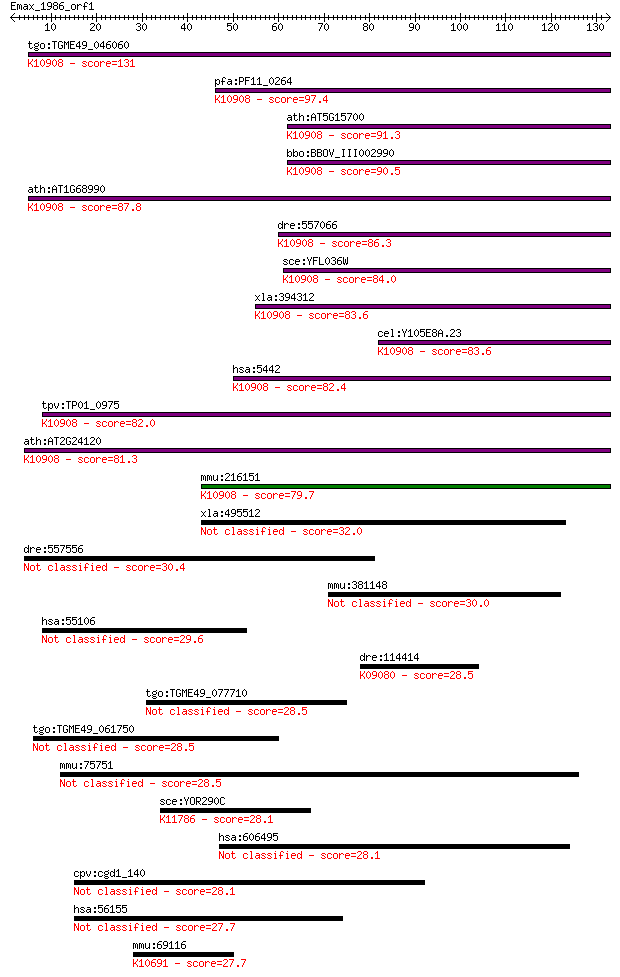
Query= Emax_1986_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_046060 DNA-dependent RNA polymerase, putative (EC:2... 131 4e-31
pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directe... 97.4 1e-20
ath:AT5G15700 DNA-directed RNA polymerase (RPOT2); K10908 DNA-... 91.3 6e-19
bbo:BBOV_III002990 17.m07284; DNA-dependent RNA polymerase; K1... 90.5 1e-18
ath:AT1G68990 DNA-directed RNA polymerase, mitochondrial (RPOM... 87.8 7e-18
dre:557066 polrmt, si:dkeyp-74c5.5, wu:fb73g10; polymerase (RN... 86.3 2e-17
sce:YFL036W RPO41; Mitochondrial RNA polymerase; single subuni... 84.0 1e-16
xla:394312 polrmt, mtRPO, polrmt-a; polymerase (RNA) mitochond... 83.6 1e-16
cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA p... 83.6 2e-16
hsa:5442 POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL; polymerase ... 82.4 4e-16
tpv:TP01_0975 DNA-directed RNA polymerase; K10908 DNA-directed... 82.0 5e-16
ath:AT2G24120 SCA3; SCA3 (SCABRA 3); DNA binding / DNA-directe... 81.3 7e-16
mmu:216151 Polrmt, 1110018N15Rik, 4932416K13, MGC118526; polym... 79.7 2e-15
xla:495512 ifit1b, ifit1l; interferon-induced protein with tet... 32.0 0.54
dre:557556 dlgap3, si:ch211-250g4.1; discs, large (Drosophila)... 30.4 1.4
mmu:381148 Gm1614; predicted gene 1614 30.0
hsa:55106 SLFN12, FLJ10260, SLFN3; schlafen family member 12 29.6
dre:114414 neurod6a, atoh2a, ndr1a, zNdr1a; neurogenic differe... 28.5 5.3
tgo:TGME49_077710 hypothetical protein 28.5 5.8
tgo:TGME49_061750 hypothetical protein 28.5 6.0
mmu:75751 Ipo4, 8430408O15Rik, AA409693, Imp4a, MGC113723, Ran... 28.5 6.1
sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit of... 28.1 6.5
hsa:606495 CYB5RL, FLJ16295; cytochrome b5 reductase-like (EC:... 28.1 7.3
cpv:cgd1_140 hypothetical protein 28.1 7.3
hsa:56155 TEX14, CT113; testis expressed 14 27.7
mmu:69116 Ubr4, 1810009A16Rik, A930005E13Rik, D930005K06Rik, G... 27.7 9.5
> tgo:TGME49_046060 DNA-dependent RNA polymerase, putative (EC:2.7.7.6);
K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=2084
Score = 131 bits (330), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 66/128 (51%), Positives = 90/128 (70%), Gaps = 8/128 (6%)
Query 5 LGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSER 64
+G++P R S + E++ Q ++ + + S TS++ S+ LRHLLQQ +++SER
Sbjct 1540 VGRLPPRDSAGLEEEIFQ-----LRRVVSNPRSSGTSAA---SIRLRHLLQQAGKLQSER 1591
Query 65 PSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGD 124
PSFLLKL+ A F A+YFPHNIDFRGRCYPLPPHLNHMGDD+ R+LL FA ++PLG
Sbjct 1592 PSFLLKLQVARCFQHLDAVYFPHNIDFRGRCYPLPPHLNHMGDDICRSLLVFAEAKPLGP 1651
Query 125 SGWRWLRI 132
G WL++
Sbjct 1652 QGLGWLKV 1659
> pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1531
Score = 97.4 bits (241), Expect = 1e-20, Method: Composition-based stats.
Identities = 43/88 (48%), Positives = 60/88 (68%), Gaps = 1/88 (1%)
Query 46 RSLLLRHLLQQETR-MKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNH 104
+ LLL+ + + + + SERP+FL KL A F + +YFPHNIDFRGR YPL PHL+H
Sbjct 1017 KYLLLKEEINRLNKCLISERPTFLQKLAVAKTFKDNDIIYFPHNIDFRGRMYPLSPHLHH 1076
Query 105 MGDDVSRALLRFARSRPLGDSGWRWLRI 132
M DD+ R+L+ FA + +G+ G WL+I
Sbjct 1077 MSDDICRSLITFAEQKEIGNKGLFWLKI 1104
> ath:AT5G15700 DNA-directed RNA polymerase (RPOT2); K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1011
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 39/71 (54%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 62 SERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRP 121
S+R LKL A A Y+PHN+DFRGR YP+PPHLNH+G D+ R +L FA RP
Sbjct 562 SQRCDTELKLSVARKMKDEEAFYYPHNMDFRGRAYPMPPHLNHLGSDLCRGVLEFAEGRP 621
Query 122 LGDSGWRWLRI 132
+G SG RWL+I
Sbjct 622 MGISGLRWLKI 632
> bbo:BBOV_III002990 17.m07284; DNA-dependent RNA polymerase;
K10908 DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1088
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/71 (54%), Positives = 49/71 (69%), Gaps = 0/71 (0%)
Query 62 SERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRP 121
SE F +L A +F + +YFP NIDFRGR YPL PHLNHM DDV R LL+FA +P
Sbjct 630 SECSLFERRLRVASDFLMEPRIYFPQNIDFRGRMYPLSPHLNHMADDVCRGLLKFADKKP 689
Query 122 LGDSGWRWLRI 132
LG+ G+ WL++
Sbjct 690 LGERGFFWLKV 700
> ath:AT1G68990 DNA-directed RNA polymerase, mitochondrial (RPOMT);
K10908 DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=976
Score = 87.8 bits (216), Expect = 7e-18, Method: Composition-based stats.
Identities = 43/128 (33%), Positives = 66/128 (51%), Gaps = 15/128 (11%)
Query 5 LGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSER 64
+G + R +PI E+ ++ + ++ + K + ++Q S+R
Sbjct 485 IGGLVDREDVPIPEEPEREDQEKFKNWRWESKKA---------------IKQNNERHSQR 529
Query 65 PSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGD 124
LKLE A Y+PHN+DFRGR YP+ P+LNH+G D+ R +L F +PLG
Sbjct 530 CDIELKLEVARKMKDEEGFYYPHNVDFRGRAYPIHPYLNHLGSDLCRGILEFCEGKPLGK 589
Query 125 SGWRWLRI 132
SG RWL+I
Sbjct 590 SGLRWLKI 597
> dre:557066 polrmt, si:dkeyp-74c5.5, wu:fb73g10; polymerase (RNA)
mitochondrial (DNA directed); K10908 DNA-directed RNA polymerase,
mitochondrial [EC:2.7.7.6]
Length=1253
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/73 (56%), Positives = 47/73 (64%), Gaps = 1/73 (1%)
Query 60 MKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARS 119
M S R L KL A N +FPHN+DFRGR YPLPP+ NH+G DV+RALL FA
Sbjct 818 MHSLRMDALYKLSIA-NHVRDKIFWFPHNMDFRGRTYPLPPYFNHLGSDVTRALLLFAEG 876
Query 120 RPLGDSGWRWLRI 132
RPLG G WL+I
Sbjct 877 RPLGPKGLDWLKI 889
> sce:YFL036W RPO41; Mitochondrial RNA polymerase; single subunit
enzyme similar to those of T3 and T7 bacteriophages; requires
a specificity subunit encoded by MTF1 for promoter recognition
(EC:2.7.7.6); K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=1351
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/72 (55%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Query 61 KSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSR 120
+S R KLE A F L LYFPHN+DFRGR YPL PH NH+G+D+SR LL F +
Sbjct 797 RSNRCDTNYKLEIARAF-LGEKLYFPHNLDFRGRAYPLSPHFNHLGNDMSRGLLIFWHGK 855
Query 121 PLGDSGWRWLRI 132
LG SG +WL+I
Sbjct 856 KLGPSGLKWLKI 867
> xla:394312 polrmt, mtRPO, polrmt-a; polymerase (RNA) mitochondrial
(DNA directed); K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=1235
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 49/78 (62%), Gaps = 1/78 (1%)
Query 55 QQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALL 114
++ + M S R L KL A + +FPHN+DFRGR YP PP+ NH+G DV+RALL
Sbjct 767 KKSSEMHSLRMDALYKLSIASHLR-DKIFWFPHNMDFRGRTYPCPPYFNHLGSDVTRALL 825
Query 115 RFARSRPLGDSGWRWLRI 132
FA RPLG G WL+I
Sbjct 826 LFAEGRPLGPKGLEWLKI 843
> cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA
polymerase, mitochondrial [EC:2.7.7.6]
Length=1138
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 31/51 (60%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 82 ALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGDSGWRWLRI 132
L+FPHN+DFRGR YPL P+L+HMGDDV+R +L+FA+S+ LG+ G+ WL++
Sbjct 704 TLFFPHNMDFRGRVYPLSPYLSHMGDDVNRCILKFAKSQKLGEKGFDWLKL 754
> hsa:5442 POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL; polymerase
(RNA) mitochondrial (DNA directed) (EC:2.7.7.6); K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1230
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 40/83 (48%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 50 LRHLLQQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDV 109
L H + M S R L +L A + + PHN+DFRGR YP PPH NH+G DV
Sbjct 762 LAHCQKVAREMHSLRAEALYRLSLAQHLR-DRVFWLPHNMDFRGRTYPCPPHFNHLGSDV 820
Query 110 SRALLRFARSRPLGDSGWRWLRI 132
+RALL FA+ RPLG G WL+I
Sbjct 821 ARALLEFAQGRPLGPHGLDWLKI 843
> tpv:TP01_0975 DNA-directed RNA polymerase; K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1225
Score = 82.0 bits (201), Expect = 5e-16, Method: Composition-based stats.
Identities = 47/125 (37%), Positives = 63/125 (50%), Gaps = 20/125 (16%)
Query 8 MPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSERPSF 67
MP + +P+ + E T I+S SS+E L L F
Sbjct 721 MPIKQHLPVNTNINLME----NINTTISNITSLSSNELSELSL----------------F 760
Query 68 LLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGDSGW 127
L +++ A FA LY P NIDFRGR YPL P+LNHM DD+ R+LL F+ R LG+ G
Sbjct 761 LRRIKIADAFANERRLYMPLNIDFRGRMYPLSPYLNHMNDDLCRSLLLFSEKRKLGNRGL 820
Query 128 RWLRI 132
WL++
Sbjct 821 FWLKV 825
> ath:AT2G24120 SCA3; SCA3 (SCABRA 3); DNA binding / DNA-directed
RNA polymerase; K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=993
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 61/129 (47%), Gaps = 15/129 (11%)
Query 4 GLGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSE 63
+ + +R +PI E+ + + Q + + ++ + E SL
Sbjct 501 NIAGLVNREDVPIPEKPSSEDPEELQSWKWSARKANKINRERHSL--------------- 545
Query 64 RPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLG 123
R LKL A Y+PHN+DFRGR YP+ PHLNH+ D+ R L FA RPLG
Sbjct 546 RCDVELKLSVARKMKDEEGFYYPHNLDFRGRAYPMHPHLNHLSSDLCRGTLEFAEGRPLG 605
Query 124 DSGWRWLRI 132
SG WL+I
Sbjct 606 KSGLHWLKI 614
> mmu:216151 Polrmt, 1110018N15Rik, 4932416K13, MGC118526; polymerase
(RNA) mitochondrial (DNA directed) (EC:2.7.7.6); K10908
DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1207
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/90 (47%), Positives = 52/90 (57%), Gaps = 2/90 (2%)
Query 43 SESRSLLLRHLLQQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHL 102
SE R L R L+ M S R L +L A + + PHN+DFRGR YP PPH
Sbjct 727 SELRKELAR-CLKVAREMHSLRSEALYRLSLAQHLR-HRVFWLPHNMDFRGRTYPCPPHF 784
Query 103 NHMGDDVSRALLRFARSRPLGDSGWRWLRI 132
NH+G D++RALL FA RPLG G WL+I
Sbjct 785 NHLGSDLARALLEFAEGRPLGPRGLDWLKI 814
> xla:495512 ifit1b, ifit1l; interferon-induced protein with tetratricopeptide
repeats 1B
Length=338
Score = 32.0 bits (71), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 41/83 (49%), Gaps = 8/83 (9%)
Query 43 SESRSLLLRHLLQQETRMK---SERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLP 99
SES S L HL + + E+ + L +LE +N L N+D RGR + +
Sbjct 2 SESLSTLKSHLEELKCHFTWGLQEKDADLEELEEKLNNQLEYL-----NMDSRGRVHNML 56
Query 100 PHLNHMGDDVSRALLRFARSRPL 122
++NH+ +D + A++ ++ +
Sbjct 57 AYVNHLKNDYAEAIVNLQKAEAI 79
> dre:557556 dlgap3, si:ch211-250g4.1; discs, large (Drosophila)
homolog-associated protein 3
Length=1059
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 40/80 (50%), Gaps = 3/80 (3%)
Query 4 GLGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISS---TSSSESRSLLLRHLLQQETRM 60
G G M +HS L+ R + GG+ GK +S S S +++L + +QE R
Sbjct 625 GPGPMKPKHSSSADNLLEGPRGSRDRVGGSLGKSASLPQNSMSLAKALTVGEEFKQEGRG 684
Query 61 KSERPSFLLKLETAVNFALS 80
+ RPS ++++++ + S
Sbjct 685 RKWRPSIAVQVDSSETLSDS 704
> mmu:381148 Gm1614; predicted gene 1614
Length=1010
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 71 LETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRP 121
++TA+ L SA+ P N+ F+ P + D +R LL A+SRP
Sbjct 249 VQTALARKLGSAVPAPSNVTFKSTAKPESTTNSQETTDSTRVLLEEAKSRP 299
> hsa:55106 SLFN12, FLJ10260, SLFN3; schlafen family member 12
Length=578
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 4/45 (8%)
Query 8 MPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRH 52
+P+ HS P+ E + QR G +G+I+ T + R L L+H
Sbjct 363 LPAPHSWPLLEW----QRQRHHCPGLSGRITYTPENLCRKLFLQH 403
> dre:114414 neurod6a, atoh2a, ndr1a, zNdr1a; neurogenic differentiation
6a; K09080 neurogenic differentiation factor 6
Length=325
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 78 ALSSALYFPHNIDFRGRCYPLPPHLN 103
A SS L+FP++I RG+ YP+ LN
Sbjct 296 AGSSDLHFPYDIHLRGQFYPVQDELN 321
> tgo:TGME49_077710 hypothetical protein
Length=556
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 31 GGTAGKISSTSSSESRSLLLRHLLQQETRMKSERPSFLLKLETA 74
G +A + ++T + E RS+L ++Q+ETR + P + + TA
Sbjct 462 GASAERTAATENPEDRSMLSLTVVQRETREEETSPRRTISVGTA 505
> tgo:TGME49_061750 hypothetical protein
Length=843
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 6/57 (10%)
Query 6 GKMPSRHSMPIAEQLQQHEAQRQQQG---GTAGKISSTSSSESRSLLLRHLLQQETR 59
G MPS+ AE Q EA+ + T GK+ + +E++S + RH+ Q+E R
Sbjct 162 GDMPSQEGATAAE-AQPQEAKESEDAFSHETTGKVET--ETENKSEVYRHMAQEELR 215
> mmu:75751 Ipo4, 8430408O15Rik, AA409693, Imp4a, MGC113723, RanBP4;
importin 4
Length=1082
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 52/116 (44%), Gaps = 17/116 (14%)
Query 12 HSMPIAEQLQQHEAQRQQQGG--TAGKISSTSSSESRSLLLRHLLQQETRMKSERPSFLL 69
H MP+ E+ + E Q++ G +S + R LL LLQ + + PS ++
Sbjct 352 HVMPMLEEALRSEDPYQRKAGFLVLAVLSDGAGDHIRQRLLYPLLQIVCK-GLDDPSQIV 410
Query 70 KLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGDS 125
+ A FAL F N+ PH++ ++V LL + +S P+G++
Sbjct 411 R--NAALFALGQ---FSENLQ---------PHISSYSEEVMPLLLSYLKSVPMGNT 452
> sce:YOR290C SNF2, GAM1, HAF1, SWI2, TYE3; Catalytic subunit
of the SWI/SNF chromatin remodeling complex involved in transcriptional
regulation; contains DNA-stimulated ATPase activity;
functions interdependently in transcriptional activation
with Snf5p and Snf6p (EC:3.6.1.-); K11786 ATP-dependent helicase
STH1/SNF2 [EC:3.6.4.-]
Length=1703
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 34 AGKISSTSSSESRSLLLRHLLQQETRMKSERPS 66
AGK + S+SE + LLR LL E + +R S
Sbjct 1238 AGKFDNKSTSEEQEALLRSLLDAEEERRKKRES 1270
> hsa:606495 CYB5RL, FLJ16295; cytochrome b5 reductase-like (EC:1.6.2.2)
Length=315
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 33/77 (42%), Gaps = 14/77 (18%)
Query 47 SLLLRHLLQQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMG 106
S+ L+ LQ++ R + R F+L E SS+ P + Y H H+G
Sbjct 224 SIYLKTFLQEQARFWNVRTFFVLSQE-------SSSEQLPWS-------YQEKTHFGHLG 269
Query 107 DDVSRALLRFARSRPLG 123
D+ + L+ R +P
Sbjct 270 QDLIKELVSCCRRKPFA 286
> cpv:cgd1_140 hypothetical protein
Length=963
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 20/78 (25%), Positives = 33/78 (42%), Gaps = 1/78 (1%)
Query 15 PIAEQLQQHEAQRQQQGGTAGKIS-STSSSESRSLLLRHLLQQETRMKSERPSFLLKLET 73
P+ + L + + Q T K S S SSE + +Q+ SE + K+E
Sbjct 241 PVEKLLDNYNKKLILQNSTGVKTSGSNRSSEKKKNDSNERNKQKLSKNSELSEYTFKIEV 300
Query 74 AVNFALSSALYFPHNIDF 91
+NF+ + L+ N F
Sbjct 301 MINFSFEAILFAIENFFF 318
> hsa:56155 TEX14, CT113; testis expressed 14
Length=1497
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 23/60 (38%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 15 PIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSL-LLRHLLQQETRMKSERPSFLLKLET 73
P E L E +QQQG + +T+S + RHL +QET K E S LL ET
Sbjct 1333 PSQELLDDIELLKQQQGSSTVLHENTASDGGGTANDQRHLEEQETDSKKEDSSMLLSKET 1392
> mmu:69116 Ubr4, 1810009A16Rik, A930005E13Rik, D930005K06Rik,
Gm1032, Gm1666, N28143, RBAF600, Zubr1, mKIAA0462, p600; ubiquitin
protein ligase E3 component n-recognin 4; K10691 E3
ubiquitin-protein ligase UBR4 [EC:6.3.2.19]
Length=5180
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 28 QQQGGTAGKISSTSSSESRSLL 49
Q+ GTAG ISSTS+S +R +L
Sbjct 3773 QEDSGTAGGISSTSASVNRYIL 3794
Lambda K H
0.320 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2099897216
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40